A High-Throughput Screen of a Library of Therapeutics Identifies Cytotoxic Substrates of P-glycoprotein
- PMID: 31515284
- PMCID: PMC6790066
- DOI: 10.1124/mol.119.115964
A High-Throughput Screen of a Library of Therapeutics Identifies Cytotoxic Substrates of P-glycoprotein
Abstract
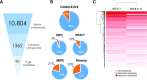
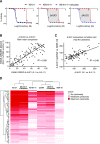
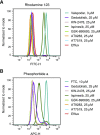
The ATP-binding cassette transporter P-glycoprotein (P-gp) is known to limit both brain penetration and oral bioavailability of many chemotherapy drugs. Although US Food and Drug Administration guidelines require that potential interactions of investigational drugs with P-gp be explored, often this information does not enter the literature. In response, we developed a high-throughput screen to identify substrates of P-gp from a series of chemical libraries, testing a total of 10,804 compounds, most of which have known mechanisms of action. We used the CellTiter-Glo viability assay to test library compounds against parental KB-3-1 human cervical adenocarcinoma cells and the colchicine-selected subline KB-8-5-11 that overexpresses P-gp. KB-8-5-11 cells were also tested in the presence of a P-gp inhibitor (tariquidar) to assess reversibility of transporter-mediated resistance. Of the tested compounds, a total of 90 P-gp substrates were identified, including 55 newly identified compounds. Substrates were confirmed using an orthogonal killing assay against human embryonic kidney-293 cells overexpressing P-gp. We confirmed that AT7159 (cyclin-dependent kinase inhibitor), AT9283, (Janus kinase 2/3 inhibitor), ispinesib (kinesin spindle protein inhibitor), gedatolisib (PKI-587, phosphoinositide 3-kinase/mammalian target of rampamycin inhibitor), GSK-690693 (AKT inhibitor), and KW-2478 (heat-shock protein 90 inhibitor) were substrates. In addition, we assessed direct ATPase stimulation. ABCG2 was also found to confer high levels of resistance to AT9283, GSK-690693, and gedatolisib, whereas ispinesib, AT7519, and KW-2478 were weaker substrates. Combinations of P-gp substrates and inhibitors were assessed to demonstrate on-target synergistic cell killing. These data identified compounds whose oral bioavailability or brain penetration may be affected by P-gp. SIGNIFICANCE STATEMENT: The ATP-binding cassette transporter P-glycoprotein (P-gp) is known to be expressed at barrier sites, where it acts to limit oral bioavailability and brain penetration of substrates. In order to identify novel compounds that are transported by P-gp, we developed a high-throughput screen using the KB-3-1 cancer cell line and its colchicine-selected subline KB-8-5-11. We screened the Mechanism Interrogation Plate (MIPE) library, the National Center for Advancing Translational Science (NCATS) pharmaceutical collection (NPC), the NCATS Pharmacologically Active Chemical Toolbox (NPACT), and a kinase inhibitor library comprising 977 compounds, for a total of 10,804 compounds. Of the 10,804 compounds screened, a total of 90 substrates were identified of which 55 were novel. P-gp expression may adversely affect the oral bioavailability or brain penetration of these compounds.
U.S. Government work not protected by U.S. copyright.
Figures



References
-
- Alvarez M, Robey R, Sandor V, Nishiyama K, Matsumoto Y, Paull K, Bates S, Fojo T. (1998) Using the national cancer institute anticancer drug screen to assess the effect of MRP expression on drug sensitivity profiles. Mol Pharmacol 54:802–814. - PubMed
-
- Ambudkar SV. (1998) Drug-stimulatable ATPase activity in crude membranes of human MDR1-transfected mammalian cells. Methods Enzymol 292:504–514. - PubMed
-
- Basseville A, Hall MD, Chau CH, Robey RW, Gottesman M, Figg WD, Bates SE. (2016) The ABCG2 multidrug transporter, in ABC Transporters - 40 Years on (George MA. ed) pp 195–226, Springer International Publishing, Cham, Switzerland.
-
- Bavetsias V, Skelton LA, Yafai F, Mitchell F, Wilson SC, Allan B, Jackman AL. (2002) The design and synthesis of water-soluble analogues of CB30865, a quinazolin-4-one-based antitumor agent. J Med Chem 45:3692–3702. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

