Transcriptomic Heterogeneity of Androgen Receptor Activity Defines a de novo low AR-Active Subclass in Treatment Naïve Primary Prostate Cancer
- PMID: 31515456
- PMCID: PMC6858964
- DOI: 10.1158/1078-0432.CCR-19-1587
Transcriptomic Heterogeneity of Androgen Receptor Activity Defines a de novo low AR-Active Subclass in Treatment Naïve Primary Prostate Cancer
Abstract
Purpose: The heterogeneity of androgen receptor (AR)-activity (AR-A) is well-characterized in heavily treated metastatic castration-resistant prostate cancer (mCRPC). However, the diversity and clinical implications of AR-A in treatment-naïve primary prostate cancer is largely unknown. We sought to characterize AR-A in localized prostate cancer and understand its molecular and clinical implications.
Experimental design: Genome-wide expression profiles from prostatectomy or biopsy samples from 19,470 patients were used, all with independent pathology review. This was comprised of prospective discovery (n = 5,239) and validation (n = 12,728) cohorts, six retrospective institutional cohorts with long-term clinical outcomes data (n = 1,170), and The Cancer Genome Atlas (n = 333).
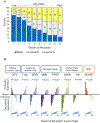
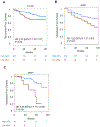
Results: A low AR-active subclass was identified, which comprised 9%-11% of each cohort, and was characterized by increased immune signaling, neuroendocrine expression, and decreased DNA repair. These tumors were predominantly ERG and basal subtype. Low AR-active tumors had significantly more rapid development of recurrence or metastatic disease across cohorts, which was maintained on multivariable analysis [HR, 2.61; 95% confidence interval (CI), 1.22-5.60; P = 0.014]. Low AR-active tumors were predicted to be more sensitive to PARP inhibition, platinum chemotherapy, and radiotherapy, and less sensitive to docetaxel and androgen-deprivation therapy. This was validated clinically, in that low AR-active tumors were less sensitive to androgen-deprivation therapy (OR, 0.41; 95% CI, 0.21-0.80; P = 0.008).
Conclusions: Leveraging large-scale transcriptomic data allowed the identification of an aggressive subtype of treatment-naïve primary prostate cancer that harbors molecular features more analogous to mCRPC. This suggests that a preexisting subgroup of patients may have tumors that are predisposed to fail multiple current standard-of-care therapies and warrant dedicated therapeutic investigation.
©2019 American Association for Cancer Research.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
Y. Liu, E. Davicioni, N. Fishbane, J. Lehrer are employees of and hold ownership interest (including patents) in Decipher Biosciences.
T. Lotan and J. Karnes received research grants from Decipher biosciences.
S.G. Zhao reports receiving commercial research support from and holds ownership interest (including patents) in Decipher Biosciences.
P.L. Nguyen reports receiving commercial research grants from Janssen, Astellas, and Bayer; holds ownership interest (including patents) in Augmenix; and is a consultant/advisory board member for Augmenix, Ferring, Blue Earth, Bayer, Cota, Dendreon, Decipher biosciences, and Nanobiotix. F.Y. Feng is an employee of PFS Genomics, and is a consultant/advisory board member for Sanofi, Janssen, Medivation/Astellas, Dandreon, Ferring, EMD Serono, Bayer, and Clovis.
Figures


Comment in
-
AR activity in treatment-naive prostate tumours.Nat Rev Urol. 2019 Nov;16(11):640. doi: 10.1038/s41585-019-0244-9. Nat Rev Urol. 2019. PMID: 31551586 No abstract available.
-
Re: Transcriptomic Heterogeneity of Androgen Receptor Activity Defines a De Novo Low AR-Active Subclass in Treatment Naïve Primary Prostate Cancer.J Urol. 2020 Mar;203(3):462. doi: 10.1097/JU.0000000000000677. Epub 2019 Dec 12. J Urol. 2020. PMID: 31829821 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

