Long noncoding RNA BFAL1 mediates enterotoxigenic Bacteroides fragilis-related carcinogenesis in colorectal cancer via the RHEB/mTOR pathway
- PMID: 31515468
- PMCID: PMC6742644
- DOI: 10.1038/s41419-019-1925-2
Long noncoding RNA BFAL1 mediates enterotoxigenic Bacteroides fragilis-related carcinogenesis in colorectal cancer via the RHEB/mTOR pathway
Abstract
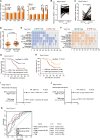
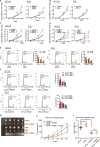
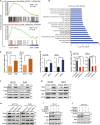
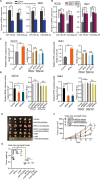
Long noncoding RNAs (lncRNAs) contribute to many steps in carcinogenesis and often serve as biomarkers or therapeutic targets for tumor diagnosis and therapy. Although the role of lncRNAs in tumor formation is becoming clear, whether lncRNAs mediate gut microbiota-induced colorectal cancer (CRC) is largely unknown. Enterotoxigenic Bacteroides fragilis (ETBF) is a well-known tumor-inducing bacterium in the human gut; however, its tumorigenic effect remains to be explored. In the present study, we revealed the mechanism by which a lncRNA participates in gut bacteria-induced carcinogenesis: Bacteroides fragilis-associated lncRNA1 (BFAL1) in CRC tissues mediates ETBF carcinogenesis. BFAL1 was highly expressed in CRC tissues compared with that in adjacent normal tissues. In vitro, BFAL1 was upregulated in ETBF-treated CRC cells. Mechanistically, ETBF promoted tumor growth via BFAL1 by activating the Ras homolog, which is the MTORC1 binding/mammalian target of the rapamycin (RHEB/mTOR) pathway. Furthermore, BFAL1 regulated RHEB expression by competitively sponging microRNAs miR-155-5p and miR-200a-3p. Clinically, both high expression of BFAL1 and high abundance of ETBF in CRC tissues predicted poor outcomes for patients with CRC. Thus, BFAL1 is a mediator of ETBF-induced carcinogenesis and may be a potential therapeutic target for ETBF-induced CRC.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Smith RA, Cokkinides V, Brooks D, Saslow D, Brawley OW. Cancer screening in the United States, 2010: a review of current American cancer society guidelines and issues in cancer screening. CA: A Cancer J. Cilni. 2010;60:99–119. - PubMed
-
- Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA: A Cancer J. Clini. 2014;64:104–117. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

