Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p
- PMID: 31515475
- PMCID: PMC6742660
- DOI: 10.1038/s41467-019-12191-9
Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p
Abstract
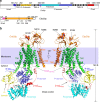
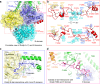
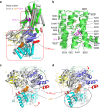
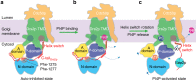
The heterodimeric eukaryotic Drs2p-Cdc50p complex is a lipid flippase that maintains cell membrane asymmetry. The enzyme complex exists in an autoinhibited form in the absence of an activator and is specifically activated by phosphatidylinositol-4-phosphate (PI4P), although the underlying mechanisms have been unclear. Here we report the cryo-EM structures of intact Drs2p-Cdc50p isolated from S. cerevisiae in apo form and in the PI4P-activated form at 2.8 Å and 3.3 Å resolution, respectively. The structures reveal that the Drs2p C-terminus lines a long groove in the cytosolic regulatory region to inhibit the flippase activity. PIP4 binding in a cytosol-proximal membrane region triggers a 90° rotation of a cytosolic helix switch that is located just upstream of the inhibitory C-terminal peptide. The rotation of the helix switch dislodges the C-terminus from the regulatory region, activating the flippase.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Palmgren Michael, Østerberg Jeppe Thulin, Nintemann Sebastian J., Poulsen Lisbeth R., López-Marqués Rosa L. Evolution and a revised nomenclature of P4 ATPases, a eukaryotic family of lipid flippases. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2019;1861(6):1135–1151. doi: 10.1016/j.bbamem.2019.02.006. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

