N-GlycositeAtlas: a database resource for mass spectrometry-based human N-linked glycoprotein and glycosylation site mapping
- PMID: 31516400
- PMCID: PMC6731604
- DOI: 10.1186/s12014-019-9254-0
N-GlycositeAtlas: a database resource for mass spectrometry-based human N-linked glycoprotein and glycosylation site mapping
Abstract
Background: N-linked glycoprotein is a highly interesting class of proteins for clinical and biological research. The large-scale characterization of N-linked glycoproteins accomplished by mass spectrometry-based glycoproteomics has provided valuable insights into the interdependence of glycoprotein structure and protein function. However, these studies focused mainly on the analysis of specific sample type, and lack the integration of glycoproteomic data from different tissues, body fluids or cell types.
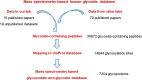
Methods: In this study, we collected the human glycosite-containing peptides identified through their de-glycosylated forms by mass spectrometry from over 100 publications and unpublished datasets generated from our laboratory. A database resource termed N-GlycositeAtlas was created and further used for the distribution analyses of glycoproteins among different human cells, tissues and body fluids. Finally, a web interface of N-GlycositeAtlas was created to maximize the utility and value of the database.
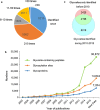
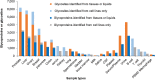
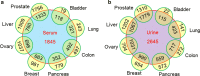
Results: The N-GlycositeAtlas database contains more than 30,000 glycosite-containing peptides (representing > 14,000 N-glycosylation sites) from more than 7200 N-glycoproteins from different biological sources including human-derived tissues, body fluids and cell lines from over 100 studies.
Conclusions: The entire human N-glycoproteome database as well as 22 sub-databases associated with individual tissues or body fluids can be downloaded from the N-GlycositeAtlas website at http://nglycositeatlas.biomarkercenter.org.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures
Similar articles
-
Mapping human N-linked glycoproteins and glycosylation sites using mass spectrometry.Trends Analyt Chem. 2019 May;114:143-150. doi: 10.1016/j.trac.2019.02.009. Epub 2019 Feb 13. Trends Analyt Chem. 2019. PMID: 31831916 Free PMC article.
-
Integrated proteomic and N-glycoproteomic analyses of doxorubicin sensitive and resistant ovarian cancer cells reveal glycoprotein alteration in protein abundance and glycosylation.Oncotarget. 2017 Feb 21;8(8):13413-13427. doi: 10.18632/oncotarget.14542. Oncotarget. 2017. PMID: 28077793 Free PMC article.
-
An integrated proteomic and glycoproteomic approach uncovers differences in glycosylation occupancy from benign and malignant epithelial ovarian tumors.Clin Proteomics. 2017 May 10;14:16. doi: 10.1186/s12014-017-9152-2. eCollection 2017. Clin Proteomics. 2017. PMID: 28491011 Free PMC article.
-
Advances in LC-MS/MS-based glycoproteomics: getting closer to system-wide site-specific mapping of the N- and O-glycoproteome.Biochim Biophys Acta. 2014 Sep;1844(9):1437-52. doi: 10.1016/j.bbapap.2014.05.002. Epub 2014 May 12. Biochim Biophys Acta. 2014. PMID: 24830338 Review.
-
Recent progress in quantitative glycoproteomics.Glycoconj J. 2012 Aug;29(5-6):249-58. doi: 10.1007/s10719-012-9398-x. Epub 2012 Jun 15. Glycoconj J. 2012. PMID: 22699565 Review.
Cited by
-
Mass spectrometry based biomarkers for early detection of HCC using a glycoproteomic approach.Adv Cancer Res. 2023;157:23-56. doi: 10.1016/bs.acr.2022.07.005. Epub 2022 Sep 6. Adv Cancer Res. 2023. PMID: 36725111 Free PMC article. Review.
-
Inter-tissue glycan heterogeneity: site-specific glycoform analysis of mouse tissue N-glycoproteomes using MS1-based glycopeptide detection method assisted by lectin microarray.Anal Bioanal Chem. 2025 Feb;417(5):973-988. doi: 10.1007/s00216-024-05686-y. Epub 2024 Dec 16. Anal Bioanal Chem. 2025. PMID: 39676134
-
Ultradeep N-glycoproteome atlas of mouse reveals spatiotemporal signatures of brain aging and neurodegenerative diseases.Nat Commun. 2025 Jul 1;16(1):5568. doi: 10.1038/s41467-025-60437-6. Nat Commun. 2025. PMID: 40593524 Free PMC article.
-
Databases and Bioinformatic Tools for Glycobiology and Glycoproteomics.Int J Mol Sci. 2020 Sep 14;21(18):6727. doi: 10.3390/ijms21186727. Int J Mol Sci. 2020. PMID: 32937895 Free PMC article. Review.
-
Quantitative analysis of fucosylated glycoproteins by immobilized lectin-affinity fluorescent labeling.RSC Adv. 2023 Feb 27;13(10):6676-6687. doi: 10.1039/d3ra00072a. eCollection 2023 Feb 21. RSC Adv. 2023. PMID: 36860533 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
