Fast and Reliable PCR Amplification from Aspergillus fumigatus Spore Suspension Without Traditional DNA Extraction
- PMID: 31518062
- PMCID: PMC6916316
- DOI: 10.1002/cpmc.89
Fast and Reliable PCR Amplification from Aspergillus fumigatus Spore Suspension Without Traditional DNA Extraction
Abstract
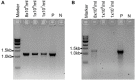
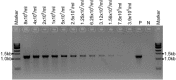
Aspergillus fumigatus is an opportunistic human pathogenic mold. DNA extraction from this fungus is usually performed by mechanical perturbation of cells, as it possesses a rigid and complex cell wall. While this is not problematic for single isolates, it can be time consuming for large numbers of strains if using traditional DNA extraction procedures. Therefore, in this article we describe a fast and efficient thermal-shock method to release DNA from spores of A. fumigatus and other filamentous fungi without the need for complex extraction methods. This is especially important for high-throughput PCR analyses of mutants in 96- or 384-well formats in a very short period of time without any concern about sample cross-contamination. This method is currently being used to validate the protein-coding gene and non-coding RNA knockout libraries in A. fumigatus generated in our laboratory, and could be used in the future for diagnostics purposes. © 2019 The Authors.
Keywords: Aspergillus fumigatus; DNA extraction; filamentous fungi; spore PCR.
© 2019 The Authors.
Figures




References
-
- Fraczek, M. G. , Bromley, M. , Buied, A. , Moore, C. B. , Rajendran, R. , Rautemaa, R. , … Bowyer, P. (2013). The cdr1B efflux transporter is associated with non‐cyp51a‐mediated itraconazole resistance in Aspergillus fumigatus . Journal of Antimicrobial Chemotherapy, 68(7), 1486–1496. doi: 10.1093/jac/dkt075 - DOI - PubMed
-
- Macdonald, D. , Thomson, D. D. , Johns, A. , Contreras Valenzuela, A. , Gilsenan, J. M. , Lord, K. M. , … Bromley, M. J. (2019). Inducible cell fusion permits use of competitive fitness profiling in the human pathogenic fungus Aspergillus fumigatus . Antimicrobial Agents and Chemotherapy, 63(1), pii: e01615–18. doi: 10.1128/AAC.01615-18 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

