When the levee breaks: a practical guide to sketching algorithms for processing the flood of genomic data
- PMID: 31519212
- PMCID: PMC6744645
- DOI: 10.1186/s13059-019-1809-x
When the levee breaks: a practical guide to sketching algorithms for processing the flood of genomic data
Abstract
Considerable advances in genomics over the past decade have resulted in vast amounts of data being generated and deposited in global archives. The growth of these archives exceeds our ability to process their content, leading to significant analysis bottlenecks. Sketching algorithms produce small, approximate summaries of data and have shown great utility in tackling this flood of genomic data, while using minimal compute resources. This article reviews the current state of the field, focusing on how the algorithms work and how genomicists can utilize them effectively. References to interactive workbooks for explaining concepts and demonstrating workflows are included at https://github.com/will-rowe/genome-sketching .
Conflict of interest statement
The author declares that he has no competing interests.
Figures
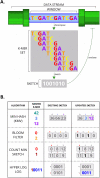
Similar articles
-
k-nonical space: sketching with reverse complements.Bioinformatics. 2024 Nov 1;40(11):btae629. doi: 10.1093/bioinformatics/btae629. Bioinformatics. 2024. PMID: 39432565 Free PMC article.
-
Sketching algorithms for genomic data analysis and querying in a secure enclave.Nat Methods. 2020 Mar;17(3):295-301. doi: 10.1038/s41592-020-0761-8. Epub 2020 Mar 4. Nat Methods. 2020. PMID: 32132732 Free PMC article.
-
RabbitSketch: a high-performance sketching library for genome analysis.Bioinformatics. 2025 May 6;41(5):btaf249. doi: 10.1093/bioinformatics/btaf249. Bioinformatics. 2025. PMID: 40286290 Free PMC article.
-
When less is more: sketching with minimizers in genomics.Genome Biol. 2024 Oct 14;25(1):270. doi: 10.1186/s13059-024-03414-4. Genome Biol. 2024. PMID: 39402664 Free PMC article. Review.
-
Prospects and limitations of full-text index structures in genome analysis.Nucleic Acids Res. 2012 Aug;40(15):6993-7015. doi: 10.1093/nar/gks408. Epub 2012 May 13. Nucleic Acids Res. 2012. PMID: 22584621 Free PMC article. Review.
Cited by
-
Sequence similarity estimation by random subsequence sketching.bioRxiv [Preprint]. 2025 May 20:2025.02.05.636706. doi: 10.1101/2025.02.05.636706. bioRxiv. 2025. PMID: 39975056 Free PMC article. Preprint.
-
Representation of k-Mer Sets Using Spectrum-Preserving String Sets.J Comput Biol. 2021 Apr;28(4):381-394. doi: 10.1089/cmb.2020.0431. Epub 2020 Dec 7. J Comput Biol. 2021. PMID: 33290137 Free PMC article.
-
To Petabytes and beyond: recent advances in probabilistic and signal processing algorithms and their application to metagenomics.Nucleic Acids Res. 2020 Jun 4;48(10):5217-5234. doi: 10.1093/nar/gkaa265. Nucleic Acids Res. 2020. PMID: 32338745 Free PMC article.
-
Simplitigs as an efficient and scalable representation of de Bruijn graphs.Genome Biol. 2021 Apr 6;22(1):96. doi: 10.1186/s13059-021-02297-z. Genome Biol. 2021. PMID: 33823902 Free PMC article.
-
k-mer approaches for biodiversity genomics.Genome Res. 2025 Feb 14;35(2):219-230. doi: 10.1101/gr.279452.124. Genome Res. 2025. PMID: 39890468 Free PMC article. Review.
References
-
- Cormode G. Data sketching. Commun ACM. 2017;60:48–55. doi: 10.1145/3080008. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

