Unique and Shared Epigenetic Programs of the CREBBP and EP300 Acetyltransferases in Germinal Center B Cells Reveal Targetable Dependencies in Lymphoma
- PMID: 31519498
- PMCID: PMC7362711
- DOI: 10.1016/j.immuni.2019.08.006
Unique and Shared Epigenetic Programs of the CREBBP and EP300 Acetyltransferases in Germinal Center B Cells Reveal Targetable Dependencies in Lymphoma
Abstract
Inactivating mutations of the CREBBP and EP300 acetyltransferases are among the most common genetic alterations in diffuse large B cell lymphoma (DLBCL) and follicular lymphoma (FL). Here, we examined the relationship between these two enzymes in germinal center (GC) B cells, the normal counterpart of FL and DLBCL, and in lymphomagenesis by using conditional GC-directed deletion mouse models targeting Crebbp or Ep300. We found that CREBBP and EP300 modulate common as well as distinct transcriptional programs implicated in separate anatomic and functional GC compartments. Consistently, deletion of Ep300 but not Crebbp impaired the fitness of GC B cells in vivo. Combined loss of Crebbp and Ep300 completely abrogated GC formation, suggesting that these proteins partially compensate for each other through common transcriptional targets. This synthetic lethal interaction was retained in CREBBP-mutant DLBCL cells and could be pharmacologically targeted with selective small molecule inhibitors of CREBBP and EP300 function. These data provide proof-of-principle for the clinical development of EP300-specific inhibitors in FL and DLBCL.
Keywords: CREBBP; EP300; acetyltransferase inhibitor; dark zone; diffuse large cell lymphoma; germinal center; light zone; synthetic lethality.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures
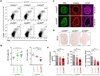

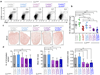



Comment in
-
Scoring a HAT-Trick against Lymphoma.Immunity. 2019 Sep 17;51(3):420-423. doi: 10.1016/j.immuni.2019.08.018. Immunity. 2019. PMID: 31533053
References
-
- Akimaru H, Chen Y, Dai P, Hou DX, Nonaka M, Smolik SM, Armstrong S, Goodman RH, and Ishii S. (1997). Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature 386, 735–738. - PubMed
-
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, and Kelly K. (1997). Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89, 1175–1184. - PubMed
-
- Bannister AJ, and Kouzarides T. (1996). The CBP co-activator is a histone acetyltransferase. Nature 384, 641–643. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

