Emergence of dominant toxigenic M1T1 Streptococcus pyogenes clone during increased scarlet fever activity in England: a population-based molecular epidemiological study
- PMID: 31519541
- PMCID: PMC6838661
- DOI: 10.1016/S1473-3099(19)30446-3
Emergence of dominant toxigenic M1T1 Streptococcus pyogenes clone during increased scarlet fever activity in England: a population-based molecular epidemiological study
Abstract
Background: Since 2014, England has seen increased scarlet fever activity unprecedented in modern times. In 2016, England's scarlet fever seasonal rise coincided with an unexpected elevation in invasive Streptococcus pyogenes infections. We describe the molecular epidemiological investigation of these events.
Methods: We analysed changes in S pyogenes emm genotypes, and notifications of scarlet fever and invasive disease in 2014-16 using regional (northwest London) and national (England and Wales) data. Genomes of 135 non-invasive and 552 invasive emm1 isolates from 2009-16 were analysed and compared with 2800 global emm1 sequences. Transcript and protein expression of streptococcal pyrogenic exotoxin A (SpeA; also known as scarlet fever or erythrogenic toxin A) in sequenced, non-invasive emm1 isolates was quantified by real-time PCR and western blot analyses.
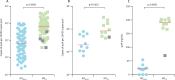
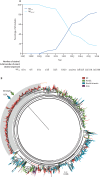
Findings: Coincident with national increases in scarlet fever and invasive disease notifications, emm1 S pyogenes upper respiratory tract isolates increased significantly in northwest London in the March to May period, from five (5%) of 96 isolates in 2014, to 28 (19%) of 147 isolates in 2015 (p=0·0021 vs 2014 values), to 47 (33%) of 144 in 2016 (p=0·0080 vs 2015 values). Similarly, invasive emm1 isolates collected nationally in the same period increased from 183 (31%) of 587 in 2015 to 267 (42%) of 637 in 2016 (p<0·0001). Sequences of emm1 isolates from 2009-16 showed emergence of a new emm1 lineage (designated M1UK)-with overlap of pharyngitis, scarlet fever, and invasive M1UK strains-which could be genotypically distinguished from pandemic emm1 isolates (M1global) by 27 single-nucleotide polymorphisms. Median SpeA protein concentration in supernatant was nine-times higher among M1UK isolates (190·2 ng/mL [IQR 168·9-200·4]; n=10) than M1global isolates (20·9 ng/mL [0·0-27·3]; n=10; p<0·0001). M1UK expanded nationally to represent 252 (84%) of all 299 emm1 genomes in 2016. Phylogenetic analysis of published datasets identified single M1UK isolates in Denmark and the USA.
Interpretation: A dominant new emm1 S pyogenes lineage characterised by increased SpeA production has emerged during increased S pyogenes activity in England. The expanded reservoir of M1UK and recognised invasive potential of emm1 S pyogenes provide plausible explanation for the increased incidence of invasive disease, and rationale for global surveillance.
Funding: UK Medical Research Council, UK National Institute for Health Research, Wellcome Trust, Rosetrees Trust, Stoneygate Trust.
Copyright © 2019 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Figures



Comment in
-
Scarlet fever changes its spots.Lancet Infect Dis. 2019 Nov;19(11):1154-1155. doi: 10.1016/S1473-3099(19)30494-3. Epub 2019 Sep 10. Lancet Infect Dis. 2019. PMID: 31519542 No abstract available.
-
Identification of Streptococcus pyogenes M1UK clone in Canada.Lancet Infect Dis. 2019 Dec;19(12):1284-1285. doi: 10.1016/S1473-3099(19)30622-X. Lancet Infect Dis. 2019. PMID: 31782392 No abstract available.
-
Against the trend: a decrease in scarlet fever in New Zealand.Lancet Infect Dis. 2019 Dec;19(12):1285-1286. doi: 10.1016/S1473-3099(19)30617-6. Lancet Infect Dis. 2019. PMID: 31782393 No abstract available.
-
8-year M type surveillance of Streptococcus pyogenes in China.Lancet Infect Dis. 2020 Jan;20(1):24-25. doi: 10.1016/S1473-3099(19)30694-2. Lancet Infect Dis. 2020. PMID: 31876494 No abstract available.
-
M1UK lineage in invasive group A streptococcus isolates from the USA.Lancet Infect Dis. 2020 May;20(5):538-539. doi: 10.1016/S1473-3099(20)30279-6. Lancet Infect Dis. 2020. PMID: 32359463 Free PMC article. No abstract available.
-
Dominance of M1UK clade among Dutch M1 Streptococcus pyogenes.Lancet Infect Dis. 2020 May;20(5):539-540. doi: 10.1016/S1473-3099(20)30278-4. Lancet Infect Dis. 2020. PMID: 32359464 No abstract available.
-
Strep A treatment, working for now.Lancet Microbe. 2023 Jan;4(1):e1. doi: 10.1016/S2666-5247(22)00360-3. Epub 2022 Dec 21. Lancet Microbe. 2023. PMID: 36565711 No abstract available.
References
-
- Krause RM. Evolving microbes and re-emerging streptococcal disease. Clin Lab Med. 2002;22:835–848. - PubMed
-
- Duncan SR, Scott S, Duncan CJ. Modelling the dynamics of scarlet fever epidemics in the 19th century. Eur J Epidemiol. 2000;16:619–626. - PubMed
-
- Lamagni T, Guy R, Chand M. Resurgence of scarlet fever in England, 2014–16: a population-based surveillance study. Lancet Infect Dis. 2018;18:180–187. - PubMed
-
- Public Health England Group A streptococcal infections: fourth update on seasonal activity, 2015/2016. May 6, 2016. https://assets.publishing.service.gov.uk/government/uploads/system/uploa...

