Ammonia Oxidizers in High-Elevation Rivers of the Qinghai-Tibet Plateau Display Distinctive Distribution Patterns
- PMID: 31519662
- PMCID: PMC6821956
- DOI: 10.1128/AEM.01701-19
Ammonia Oxidizers in High-Elevation Rivers of the Qinghai-Tibet Plateau Display Distinctive Distribution Patterns
Abstract
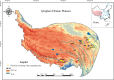
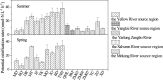
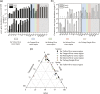
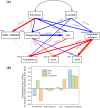
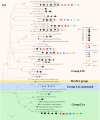
Ammonia-oxidizing bacteria (AOB) and archaea (AOA) as well as comammox catalyze ammonia oxidation. The distribution and biogeography of these ammonia oxidizers might be distinctive in high-elevation rivers, which are generally characterized by low temperature and low ammonium concentration but strong solar radiation; however, these characteristics have rarely been documented. This study explored the abundance, community, and activity of ammonia oxidizers in the overlying water of five rivers in the Qinghai-Tibet Plateau (QTP). Potential nitrification rates in these rivers ranged from 5.4 to 38.4 nmol N liter-1 h-1, and they were significantly correlated with ammonium concentration rather than temperature. Comammox were found in 25 of the total 28 samples, and they outnumbered AOA in three samples. Contrary to most studied low-elevation rivers, average AOB amoA gene abundance was significantly higher than that of AOA, and AOB/AOA ratios increased with decreasing water temperature. The Simpson index of the AOA community increased with elevation (P < 0.05), and AOA and AOB communities exhibited high dissimilarities with low-elevation rivers. Cold-adapted (Nitrosospira amoA cluster 1, 33.6%) and oligotrophic (Nitrosomonas amoA cluster 6a, 31.7%) groups accounted for large proportions in the AOB community. Suspended sediment concentration exerted significant effects on ammonia oxidizer abundance (r > 0.56), and owing to their elevational variations in source and concentration, suspended sediments facilitated distance-decay patterns for AOA and AOB community similarities. This study demonstrates distinctive biogeography and distribution patterns for ammonia oxidizers in high-elevation rivers of the QTP. Extensive research should be conducted to explore the role of these microbes in the nitrogen cycle of this zone.IMPORTANCE Ammonia-oxidizing archaea (AOA) and bacteria (AOB) as well as comammox contribute to ammonia oxidation, which plays significant roles in riverine nitrogen cycle and N2O production. Source regions of numerous rivers in the world lie in high-elevation zones, but the abundance, community, and activity of ammonia oxidizers in rivers in high-elevation regions have rarely been investigated. This study revealed distinctive distribution patterns and community structures for ammonia oxidizers in five high-elevation rivers of the Qinghai-Tibet Plateau, and the individual and combined effects of low temperature, low nutrients, and strong solar radiation on ammonia oxidizers were elucidated. The findings of this study are helpful to broaden our knowledge on the biogeography and distribution pattern of ammonia oxidizers in river systems. Moreover, this study provides some implications to predict the performance of ammonia oxidizers in high-elevation rivers and its variations under global climate warming.
Keywords: Qinghai-Tibet Plateau; ammonia-oxidizing archaea (AOA); comammox Nitrospira; distance-decay pattern; high-elevation rivers; suspended sediment.
Copyright © 2019 American Society for Microbiology.
Figures




References
-
- Beaulieu JJ, Tank JL, Hamilton SK, Wollheim WM, Hall RO, Mulholland PJ, Peterson BJ, Ashkenas LR, Cooper LW, Dahm CN, Dodds WK, Grimm NB, Johnson SL, McDowell WH, Poole GC, Valett HM, Arango CP, Bernot MJ, Burgin AJ, Crenshaw CL, Helton AM, Johnson LT, O'Brien JM, Potter JD, Sheibley RW, Sobota DJ, Thomas SM. 2011. Nitrous oxide emission from denitrification in stream and river networks. Proc Natl Acad Sci USA 108:214–219. doi:10.1073/pnas.1011464108. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

