Experimental models and tools to tackle glioblastoma
- PMID: 31519690
- PMCID: PMC6765190
- DOI: 10.1242/dmm.040386
Experimental models and tools to tackle glioblastoma
Abstract
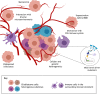
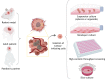
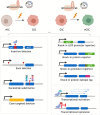
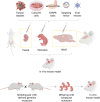
Glioblastoma multiforme (GBM) is one of the deadliest human cancers. Despite increasing knowledge of the genetic and epigenetic changes that underlie tumour initiation and growth, the prognosis for GBM patients remains dismal. Genome analysis has failed to lead to success in the clinic. Fresh approaches are needed that can stimulate new discoveries across all levels: cell-intrinsic mechanisms (transcriptional/epigenetic and metabolic), cell-cell signalling, niche and microenvironment, systemic signals, immune regulation, and tissue-level physical forces. GBMs are inherently extremely challenging: tumour detection occurs too late, and cells infiltrate widely, hiding in quiescent states behind the blood-brain barrier. The complexity of the brain tissue also provides varied and complex microenvironments that direct cancer cell fates. Phenotypic heterogeneity is therefore superimposed onto pervasive genetic heterogeneity. Despite this bleak outlook, there are reasons for optimism. A myriad of complementary, and increasingly sophisticated, experimental approaches can now be used across the research pipeline, from simple reductionist models devised to delineate molecular and cellular mechanisms, to complex animal models required for preclinical testing of new therapeutic approaches. No single model can cover the breadth of unresolved questions. This Review therefore aims to guide investigators in choosing the right model for their question. We also discuss the recent convergence of two key technologies: human stem cell and cancer stem cell culture, as well as CRISPR/Cas tools for precise genome manipulations. New functional genetic approaches in tailored models will likely fuel new discoveries, new target identification and new therapeutic strategies to tackle GBM.
Keywords: Brain tumour; CRISPR/Cas9; Cancer; Central nervous system; GBM; Human; In vitro; Mouse; Xenograft.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures
References
-
- Alcantara Llaguno S., Chen J., Kwon C.-H., Jackson E. L., Li Y., Burns D. K., Alvarez-Buylla A. and Parada L. F. (2009). Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 15, 45-56. 10.1016/j.ccr.2008.12.006 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

