Layer 4 of mouse neocortex differs in cell types and circuit organization between sensory areas
- PMID: 31519874
- PMCID: PMC6744474
- DOI: 10.1038/s41467-019-12058-z
Layer 4 of mouse neocortex differs in cell types and circuit organization between sensory areas
Erratum in
-
Author Correction: Layer 4 of mouse neocortex differs in cell types and circuit organization between sensory areas.Nat Commun. 2019 Nov 4;10(1):5075. doi: 10.1038/s41467-019-12769-3. Nat Commun. 2019. PMID: 31685814 Free PMC article.
Abstract
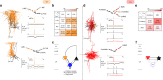
Layer 4 (L4) of mammalian neocortex plays a crucial role in cortical information processing, yet a complete census of its cell types and connectivity remains elusive. Using whole-cell recordings with morphological recovery, we identified one major excitatory and seven inhibitory types of neurons in L4 of adult mouse visual cortex (V1). Nearly all excitatory neurons were pyramidal and all somatostatin-positive (SOM+) non-fast-spiking interneurons were Martinotti cells. In contrast, in somatosensory cortex (S1), excitatory neurons were mostly stellate and SOM+ interneurons were non-Martinotti. These morphologically distinct SOM+ interneurons corresponded to different transcriptomic cell types and were differentially integrated into the local circuit with only S1 neurons receiving local excitatory input. We propose that cell type specific circuit motifs, such as the Martinotti/pyramidal and non-Martinotti/stellate pairs, are used across the cortex as building blocks to assemble cortical circuits.
Conflict of interest statement
The authors declare no competing interests.
Figures

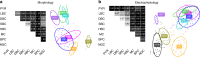


References
-
- Simons DJ, Woolsey TA. Morphology of Golgi-Cox-impregnated barrel neurons in rat SmI cortex. J. Comp. Neurol. 1984;230:119–132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

