UPF1/SMG7-dependent microRNA-mediated gene regulation
- PMID: 31519907
- PMCID: PMC6744440
- DOI: 10.1038/s41467-019-12123-7
UPF1/SMG7-dependent microRNA-mediated gene regulation
Abstract
The stability and quality of metazoan mRNAs are under microRNA (miRNA)-mediated and nonsense-mediated control. Although UPF1, a core mediator of nonsense-mediated mRNA decay (NMD), mediates the decay of target mRNA in a 3'UTR-length-dependent manner, the detailed mechanism remains unclear. Here, we suggest that 3'UTR-length-dependent mRNA decay is not mediated by nonsense mRNAs but rather by miRNAs that downregulate target mRNAs via Ago-associated UPF1/SMG7. Global analyses of mRNAs in response to UPF1 RNA interference in miRNA-deficient cells reveal that 3'UTR-length-dependent mRNA decay by UPF1 requires canonical miRNA targeting. The destabilization of miRNA targets is accomplished by the combination of Ago2 and UPF1/SMG7, which may recruit the CCR4-NOT deadenylase complex. Indeed, loss of the SMG7-deadenylase complex interaction increases the levels of transcripts regulated by UPF1-SMG7. This UPF1/SMG7-dependent miRNA-mediated mRNA decay pathway may enable miRNA targeting to become more predictable and expand the miRNA-mRNA regulatory network.
Conflict of interest statement
The authors declare no competing interests.
Figures

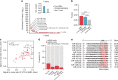
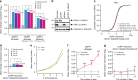
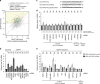
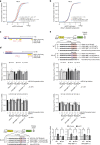

Similar articles
-
Dissecting the functions of SMG5, SMG7, and PNRC2 in nonsense-mediated mRNA decay of human cells.RNA. 2018 Apr;24(4):557-573. doi: 10.1261/rna.063719.117. Epub 2018 Jan 18. RNA. 2018. PMID: 29348139 Free PMC article.
-
A post-translational regulatory switch on UPF1 controls targeted mRNA degradation.Genes Dev. 2014 Sep 1;28(17):1900-16. doi: 10.1101/gad.245506.114. Genes Dev. 2014. PMID: 25184677 Free PMC article.
-
Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways.RNA. 2017 Feb;23(2):189-201. doi: 10.1261/rna.059055.116. Epub 2016 Nov 18. RNA. 2017. PMID: 27864472 Free PMC article.
-
UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond.RNA. 2019 Apr;25(4):407-422. doi: 10.1261/rna.070136.118. Epub 2019 Jan 17. RNA. 2019. PMID: 30655309 Free PMC article. Review.
-
Up-frameshift protein 1 (UPF1): multitalented entertainer in RNA decay.Drug Discov Ther. 2012 Apr;6(2):55-61. Drug Discov Ther. 2012. PMID: 22622014 Review.
Cited by
-
Translation-dependent and -independent mRNA decay occur through mutually exclusive pathways defined by ribosome density during T cell activation.Genome Res. 2024 Apr 25;34(3):394-409. doi: 10.1101/gr.277863.123. Genome Res. 2024. PMID: 38508694 Free PMC article.
-
Inferring gene regulatory networks with graph convolutional network based on causal feature reconstruction.Sci Rep. 2024 Sep 12;14(1):21342. doi: 10.1038/s41598-024-71864-8. Sci Rep. 2024. PMID: 39266676 Free PMC article.
-
RNA-biology ruling cancer progression? Focus on 3'UTRs and splicing.Cancer Metastasis Rev. 2020 Sep;39(3):887-901. doi: 10.1007/s10555-020-09884-9. Cancer Metastasis Rev. 2020. PMID: 32361913 Review.
-
Analysis of recent shared ancestry in a familial cohort identifies coding and noncoding autism spectrum disorder variants.NPJ Genom Med. 2022 Feb 21;7(1):13. doi: 10.1038/s41525-022-00284-2. NPJ Genom Med. 2022. PMID: 35190550 Free PMC article.
-
ZNF740 facilitates the malignant progression of hepatocellular carcinoma via the METTL3/HIF‑1A signaling axis.Int J Oncol. 2024 Nov;65(5):105. doi: 10.3892/ijo.2024.5693. Epub 2024 Sep 20. Int J Oncol. 2024. PMID: 39301659 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

