Selective autophagy maintains centrosome integrity and accurate mitosis by turnover of centriolar satellites
- PMID: 31519908
- PMCID: PMC6744468
- DOI: 10.1038/s41467-019-12094-9
Selective autophagy maintains centrosome integrity and accurate mitosis by turnover of centriolar satellites
Abstract
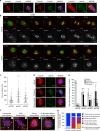
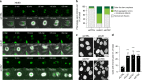
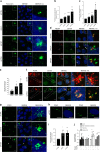
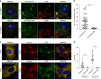
The centrosome is the master orchestrator of mitotic spindle formation and chromosome segregation in animal cells. Centrosome abnormalities are frequently observed in cancer, but little is known of their origin and about pathways affecting centrosome homeostasis. Here we show that autophagy preserves centrosome organization and stability through selective turnover of centriolar satellite components, a process we termed doryphagy. Autophagy targets the satellite organizer PCM1 by interacting with GABARAPs via a C-terminal LIR motif. Accordingly, autophagy deficiency results in accumulation of large abnormal centriolar satellites and a resultant dysregulation of centrosome composition. These alterations have critical impact on centrosome stability and lead to mitotic centrosome fragmentation and unbalanced chromosome segregation. Our findings identify doryphagy as an important centrosome-regulating pathway and bring mechanistic insights to the link between autophagy dysfunction and chromosomal instability. In addition, we highlight the vital role of centriolar satellites in maintaining centrosome integrity.
Conflict of interest statement
The authors declare no competing interests.
Figures