B cells sustain inflammation and predict response to immune checkpoint blockade in human melanoma
- PMID: 31519915
- PMCID: PMC6744450
- DOI: 10.1038/s41467-019-12160-2
B cells sustain inflammation and predict response to immune checkpoint blockade in human melanoma
Abstract
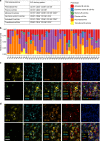
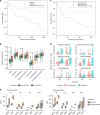
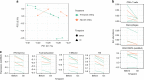
Tumor associated inflammation predicts response to immune checkpoint blockade in human melanoma. Current theories on regulation of inflammation center on anti-tumor T cell responses. Here we show that tumor associated B cells are vital to melanoma associated inflammation. Human B cells express pro- and anti-inflammatory factors and differentiate into plasmablast-like cells when exposed to autologous melanoma secretomes in vitro. This plasmablast-like phenotype can be reconciled in human melanomas where plasmablast-like cells also express T cell-recruiting chemokines CCL3, CCL4, CCL5. Depletion of B cells in melanoma patients by anti-CD20 immunotherapy decreases tumor associated inflammation and CD8+ T cell numbers. Plasmablast-like cells also increase PD-1+ T cell activation through anti-PD-1 blockade in vitro and their frequency in pretherapy melanomas predicts response and survival to immune checkpoint blockade. Tumor associated B cells therefore orchestrate and sustain melanoma inflammation and may represent a predictor for survival and response to immune checkpoint blockade therapy.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Ascierto Paolo A., Long Georgina V., Robert Caroline, Brady Benjamin, Dutriaux Caroline, Di Giacomo Anna Maria, Mortier Laurent, Hassel Jessica C., Rutkowski Piotr, McNeil Catriona, Kalinka-Warzocha Ewa, Savage Kerry J., Hernberg Micaela M., Lebbé Celeste, Charles Julie, Mihalcioiu Catalin, Chiarion-Sileni Vanna, Mauch Cornelia, Cognetti Francesco, Ny Lars, Arance Ana, Svane Inge Marie, Schadendorf Dirk, Gogas Helen, Saci Abdel, Jiang Joel, Rizzo Jasmine, Atkinson Victoria. Survival Outcomes in Patients With Previously Untreated BRAF Wild-Type Advanced Melanoma Treated With Nivolumab Therapy. JAMA Oncology. 2019;5(2):187. doi: 10.1001/jamaoncol.2018.4514. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

