Cancer biology as revealed by the research autopsy
- PMID: 31519982
- PMCID: PMC7453489
- DOI: 10.1038/s41568-019-0199-4
Cancer biology as revealed by the research autopsy
Abstract
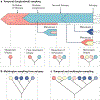
A research autopsy is a post-mortem medical procedure performed on a deceased individual with the primary goal of collecting tissue to support basic and translational research. This approach has increasingly been used to investigate the pathophysiological mechanisms of cancer evolution, metastasis and treatment resistance. In this Review, we discuss the rationale for the use of research autopsies in cancer research and provide an evidence-based discussion of the quality of post-mortem tissues compared with other types of biospecimens. We also discuss the advantages of using post-mortem tissues over other types of biospecimens, including the large amounts of tissue that can be obtained and the extent of multiregion sampling that is achievable, which is not otherwise possible in living patients. We highlight how the research autopsy has supported the identification of the clonal origins and modes of spread among metastases, the extent that selective pressures imposed by treatments cause bottlenecks leading to parallel and convergent tumour evolution, and the creation of rare tissue banks and patient-derived model systems. Finally, we comment on the future of the research autopsy as an integral component of precision medicine strategies.
Conflict of interest statement
Competing interests
C.I.A.-D. and T.J.H. have received research support from Bristol-Myers Squibb. The other authors declare no competing interests.
Figures


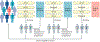

References
-
- Blokker BM et al. Conventional autopsy versus minimally invasive autopsy with postmortem MRI, CT, and CT-guided biopsy: comparison of diagnostic performance. Radiology 289, 658–667 (2018). - PubMed
-
- Kretzschmar H Brain banking: opportunities, challenges and meaning for the future. Nat Rev. Neurosci 10, 70–78 (2009). - PubMed
-
- Hajdu SI A note from history: the first printed case reports of cancer. Cancer 116, 2493–2498 (2010). - PubMed
-
- Mariette C et al. Consensus on the pathological definition and classification of poorly cohesive gastric carcinoma. Gastric Cancer 22, 1–9 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

