MicroRNA Profiling Reveals Distinct Signatures in Degenerative Rotator Cuff Pathologies
- PMID: 31520478
- PMCID: PMC6973295
- DOI: 10.1002/jor.24473
MicroRNA Profiling Reveals Distinct Signatures in Degenerative Rotator Cuff Pathologies
Abstract
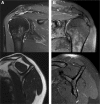
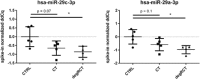
MicroRNAs (miRNAs) have emerged as key regulators orchestrating a wide range of inflammatory and fibrotic diseases. However, the role of miRNAs in degenerative shoulder joint disorders is poorly understood. The aim of this explorative case-control study was to identify pathology-related, circulating miRNAs in patients with chronic rotator cuff tendinopathy and degenerative rotator cuff tears (RCT). In 2017, 15 patients were prospectively enrolled and assigned to three groups based on the diagnosed pathology: (i) no shoulder pathology, (ii) chronic rotator cuff tendinopathy, and (iii) degenerative RCTs. In total, 14 patients were included. Venous blood samples ("liquid biopsies") were collected from each patient and serum levels of 187 miRNAs were determined. Subsequently, the change in expression of nine candidate miRNAs was verified in tendon biopsy samples, collected from patients who underwent arthroscopic shoulder surgery between 2015 and 2018. Overall, we identified several miRNAs to be progressively deregulated in sera from patients with either chronic rotator cuff tendinopathy or degenerative RCTs. Importantly, for the several of these miRNAs candidates repression was also evident in tendon biopsies harvested from patients who were treated for a supraspinatus tendon tear. As similar expression profiles were determined for tendon samples, the newly identified systemic miRNA signature has potential as novel diagnostic or prognostic biomarkers for degenerative rotator cuff pathologies. © 2019 The Authors. Journal of Orthopaedic Research® published by Wiley Periodicals, Inc. on behalf of Orthopaedic Research Society. Inc. J Orthop Res 38:202-211, 2020.
Keywords: biomarker; circulating microRNA; rotator cuff; tendinopathy; tendon degeneration.
© 2019 The Authors. Journal of Orthopaedic Research® published by Wiley Periodicals, Inc. on behalf of Orthopaedic Research Society. Inc.
Figures




Similar articles
-
Risk Factors for Rotator Cuff Disease: An Experimental Study on Intact Human Subscapularis Tendons.J Orthop Res. 2020 Jan;38(1):182-191. doi: 10.1002/jor.24385. Epub 2019 Jun 25. J Orthop Res. 2020. PMID: 31161610 Free PMC article.
-
Evaluating the role of subacromial impingement in rotator cuff tendinopathy: Development and analysis of a novel murine model.J Orthop Res. 2018 Oct;36(10):2780-2788. doi: 10.1002/jor.24026. Epub 2018 May 24. J Orthop Res. 2018. PMID: 29683224 Free PMC article.
-
Degree of tendon degeneration and stage of rotator cuff disease.Knee Surg Sports Traumatol Arthrosc. 2017 Jul;25(7):2100-2108. doi: 10.1007/s00167-016-4376-7. Epub 2016 Nov 28. Knee Surg Sports Traumatol Arthrosc. 2017. PMID: 27896393
-
US of the shoulder: rotator cuff and non-rotator cuff disorders.Radiographics. 2006 Jan-Feb;26(1):e23. doi: 10.1148/rg.e23. Radiographics. 2006. PMID: 16352733 Review.
-
Shoulder MRI refinements: differentiation of rotator cuff tear from artifacts and tendonosis, and reassessment of normal findings.Semin Ultrasound CT MR. 2001 Aug;22(4):383-95. doi: 10.1016/s0887-2171(01)90028-9. Semin Ultrasound CT MR. 2001. PMID: 11513161 Review.
Cited by
-
Systematic identification of aberrant non-coding RNAs and their mediated modules in rotator cuff tears.Front Mol Biosci. 2022 Aug 30;9:940290. doi: 10.3389/fmolb.2022.940290. eCollection 2022. Front Mol Biosci. 2022. PMID: 36111133 Free PMC article.
-
Long noncoding RNA H19 accelerates tenogenic differentiation by modulating miR-140-5p/VEGFA signaling.Eur J Histochem. 2021 Sep 7;65(3):3297. doi: 10.4081/ejh.2021.3297. Eur J Histochem. 2021. PMID: 34494412 Free PMC article.
-
Rotator Cuff Health, Pathology, and Repair in the Perspective of Hyperlipidemia.J Orthop Sports Med. 2022;4(4):263-275. doi: 10.26502/josm.511500063. Epub 2022 Oct 17. J Orthop Sports Med. 2022. PMID: 36381991 Free PMC article.
-
Biological and Mechanical Factors and Epigenetic Regulation Involved in Tendon Healing.Stem Cells Int. 2023 Jan 9;2023:4387630. doi: 10.1155/2023/4387630. eCollection 2023. Stem Cells Int. 2023. PMID: 36655033 Free PMC article. Review.
-
Computational Analysis of miR-140 and miR-135 as Potential Targets to Develop Combinatorial Therapeutics for Degenerative Tendinopathy.Clin Orthop Surg. 2023 Jun;15(3):463-476. doi: 10.4055/cios22237. Epub 2023 May 15. Clin Orthop Surg. 2023. PMID: 37274502 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

