Transcriptional regulation of stilbene synthases in grapevine germplasm differentially susceptible to downy mildew
- PMID: 31521112
- PMCID: PMC6744718
- DOI: 10.1186/s12870-019-2014-5
Transcriptional regulation of stilbene synthases in grapevine germplasm differentially susceptible to downy mildew
Abstract
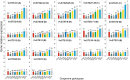
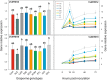
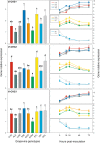
Background: To limit the impact of the downy mildew disease of grapevine and reduce the need to recur to chemical treatments, an effective strategy might be recovering adaptive resistance traits in both cultivated and wild V. vinifera germplasm. Considering that stilbenes represent the most important class of phytoalexins in the Vitaceae, the constitutive expression and transcriptional activation of all the functional members of the stilbene synthase gene family were analysed in a group of nine grapevine genotypes following artificial infection with the oomycete Plasmopara viticola, the causal agent of the disease. In addition, in the same genotypes we analyzed the expression of genes encoding for two transcription factors involved in the transcriptional regulation of the stilbene synthase genes, namely VvMYB14 and VvMYB15, and of genes encoding for chalcone synthases.
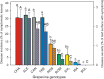
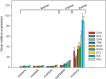
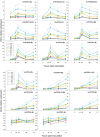
Results: Downy mildew incidence and severity ranged from nihil to high in the grapevine genotypes considered, being low to moderate in a subgroup of V. vinifera genotypes. The constitutive expression of the stilbene synthase genes as well as the extent of their transcriptional activation following P. viticola inoculation appeared to be inversely related to the proneness to develop disease symptoms upon infection. In a specular manner, following P. viticola inoculation all the chalcone synthase genes were up-regulated in the susceptible grapevine genotypes and down-regulated in the resistant ones. The infection brought by P. viticola appeared to elicit a co-ordinated and sequential transcriptional activation of distinct stilbene synthase genes subsets, each of which may be regulated by a distinct and specific MYB transcription factor.
Conclusions: The present results suggest that the induction of stilbene biosynthesis may contribute to the basal immunity against the downy mildew of grapevine, thus representing an adaptive resistance trait to recover, in both cultivated and wild V. vinifera germplasm. During the early stages of P. viticola infection, an antagonistic interaction between flavonol and stilbene biosynthesis might occur, whose outcome might determine the subsequent extent of disease symptoms. Further studies are needed to decipher the possible regulatory mechanisms involved in the antagonistic crosstalk between these two metabolic pathways in resistant and susceptible genotypes in response to P. viticola.
Keywords: Chalcone synthase; Downy mildew susceptibility; Grapevine germplasm; MYB14 and MYB15; Plant-pathogen interactions; Plasmopara viticola; Stilbene synthase; Vitis vinifera L.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Resistance to Plasmopara viticola in a grapevine segregating population is associated with stilbenoid accumulation and with specific host transcriptional responses.BMC Plant Biol. 2011 Aug 12;11:114. doi: 10.1186/1471-2229-11-114. BMC Plant Biol. 2011. PMID: 21838877 Free PMC article.
-
Genetic diversity of stilbene metabolism in Vitis sylvestris.J Exp Bot. 2015 Jun;66(11):3243-57. doi: 10.1093/jxb/erv137. Epub 2015 Apr 6. J Exp Bot. 2015. PMID: 25873669 Free PMC article.
-
General and species-specific transcriptional responses to downy mildew infection in a susceptible (Vitis vinifera) and a resistant (V. riparia) grapevine species.BMC Genomics. 2010 Feb 18;11:117. doi: 10.1186/1471-2164-11-117. BMC Genomics. 2010. PMID: 20167053 Free PMC article.
-
The pathogenicity of Plasmopara viticola: a review of evolutionary dynamics, infection strategies and effector molecules.BMC Plant Biol. 2024 Apr 24;24(1):327. doi: 10.1186/s12870-024-05037-0. BMC Plant Biol. 2024. PMID: 38658826 Free PMC article. Review.
-
Plasmopara viticola the Causal Agent of Downy Mildew of Grapevine: From Its Taxonomy to Disease Management.Front Microbiol. 2022 May 11;13:889472. doi: 10.3389/fmicb.2022.889472. eCollection 2022. Front Microbiol. 2022. PMID: 35633680 Free PMC article. Review.
Cited by
-
Leaf Polyphenolic Profile as a Determinant of Croatian Native Grapevine Varieties' Susceptibility to Plasmopara viticola.Front Plant Sci. 2022 Mar 11;13:836318. doi: 10.3389/fpls.2022.836318. eCollection 2022. Front Plant Sci. 2022. PMID: 35360327 Free PMC article.
-
Resveratrol: Its Path from Isolation to Therapeutic Action in Eye Diseases.Antioxidants (Basel). 2022 Dec 12;11(12):2447. doi: 10.3390/antiox11122447. Antioxidants (Basel). 2022. PMID: 36552655 Free PMC article. Review.
-
Short-Term Gaseous Treatments Improve Rachis Browning in Red and White Table Grapes Stored at Low Temperature: Molecular Mechanisms Underlying Its Beneficial Effect.Int J Mol Sci. 2022 Nov 1;23(21):13304. doi: 10.3390/ijms232113304. Int J Mol Sci. 2022. PMID: 36362091 Free PMC article.
-
Constitutive expression of VviNAC17 transcription factor significantly induces the synthesis of flavonoids and other phenolics in transgenic grape berry cells.Front Plant Sci. 2022 Jul 29;13:964621. doi: 10.3389/fpls.2022.964621. eCollection 2022. Front Plant Sci. 2022. PMID: 35968093 Free PMC article.
-
Hybrid Vitis Cultivars with American or Asian Ancestries Show Higher Tolerance towards Grapevine Trunk Diseases.Plants (Basel). 2023 Jun 15;12(12):2328. doi: 10.3390/plants12122328. Plants (Basel). 2023. PMID: 37375953 Free PMC article.
References
-
- OIV . International Organization of Vine and Wine, Statistical report on world vitiviniculture. 2018.
-
- Galet P. Les Maladies et les Parasites de la Vigne, Vol 1. Montpellier: Imp. Paysan du Midi; 1997.
-
- Muganu M, Balestra GM, Magro P, Pettinari G, Bignami C. Susceptibility of local grape cultivars to Plasmopara viticola and response to copper compounds with low cupric salts concentration in Latium (Central Italy) Acta Hortic. 2007;754:373–378. doi: 10.17660/ActaHortic.2007.754.49. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

