Structural insights into coronavirus entry
- PMID: 31522710
- PMCID: PMC7112261
- DOI: 10.1016/bs.aivir.2019.08.002
Structural insights into coronavirus entry
Abstract
Coronaviruses (CoVs) have caused outbreaks of deadly pneumonia in humans since the beginning of the 21st century. The severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in 2002 and was responsible for an epidemic that spread to five continents with a fatality rate of 10% before being contained in 2003 (with additional cases reported in 2004). The Middle-East respiratory syndrome coronavirus (MERS-CoV) emerged in the Arabian Peninsula in 2012 and has caused recurrent outbreaks in humans with a fatality rate of 35%. SARS-CoV and MERS-CoV are zoonotic viruses that crossed the species barrier using bats/palm civets and dromedary camels, respectively. No specific treatments or vaccines have been approved against any of the six human coronaviruses, highlighting the need to investigate the principles governing viral entry and cross-species transmission as well as to prepare for zoonotic outbreaks which are likely to occur due to the large reservoir of CoVs found in mammals and birds. Here, we review our understanding of the infection mechanism used by coronaviruses derived from recent structural and biochemical studies.
Keywords: Coronavirus; Fusion protein; Membrane fusion; Proteolytic activation; Spike glycoprotein; Vaccine design.
© 2019 Elsevier Inc. All rights reserved.
Figures

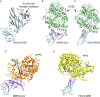
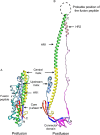
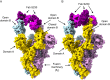

References
-
- Bakkers M.J., Lang Y., Feitsma L.J., Hulswit R.J., de Poot S.A., van Vliet A.L., Margine I., de Groot-Mijnes J.D., van Kuppeveld F.J., Langereis M.A., Huizinga E.G., de Groot R.J. Betacoronavirus adaptation to humans involved progressive loss of hemagglutinin-esterase lectin activity. Cell Host Microbe. 2017;21:356. - PMC - PubMed
-
- Bakkers M.J., Zeng Q., Feitsma L.J., Hulswit R.J., Li Z., Westerbeke A., van Kuppeveld F.J., Boons G.J., Langereis M.A., Huizinga E.G., de Groot R.J. Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation. Proc. Natl. Acad. Sci. U. S. A. 2016;113 - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

