Chronic Inflammation Directs an Olfactory Stem Cell Functional Switch from Neuroregeneration to Immune Defense
- PMID: 31523027
- PMCID: PMC6778045
- DOI: 10.1016/j.stem.2019.08.011
Chronic Inflammation Directs an Olfactory Stem Cell Functional Switch from Neuroregeneration to Immune Defense
Abstract
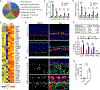
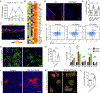
Although olfactory mucosa possesses long-lived horizontal basal stem cells (HBCs) and remarkable regenerative capacity, the function of human olfactory neuroepithelium is significantly impaired in chronic inflammatory rhinosinusitis. Here, we show that, while inflammation initially damages olfactory neurons and activates HBC-mediated regeneration, continued inflammation locks HBCs in an undifferentiated state. Global gene expression in mouse HBCs reveals broad upregulation of NF-κB-regulated cytokines and chemokines including CCL19, CCL20, and CXCL10, accompanied by enhancement of "stemness"-related transcription factors. Loss-of-function studies identify an NF-κB-dependent role of HBCs in amplifying inflammatory signaling, contributing to macrophage and T cell local proliferation. Chronically activated HBCs signal macrophages to maintain immune defense and prevent Treg development. In diseased human olfactory tissue, activated HBCs in a P63+ undifferentiated state similarly contribute to inflammation through chemokine production. These observations establish a mechanism of chronic rhinosinusitis-associated olfactory loss, caused by a functional switch of neuroepithelial stem cells from regeneration to immune defense.
Keywords: NF-kB; basal stem cell; chemokine; chronic inflammation; neural stem cell; neurogenesis; olfactory epithelium; regeneration.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures




Comment in
-
Smelling Danger: Olfactory Stem Cells Control Immune Defense during Chronic Inflammation.Cell Stem Cell. 2019 Oct 3;25(4):449-451. doi: 10.1016/j.stem.2019.09.006. Cell Stem Cell. 2019. PMID: 31585089
References
-
- Afonina IS, Zhong Z, Karin M, and Beyaert R (2017). Limiting inflammation-the negative regulation of NF-kappaB and the NLRP3 inflammasome. Nat Immunol 18, 861–869. - PubMed
-
- Artis D (2008). Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol 8, 411–420. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

