Identification of core genes and pathways in type 2 diabetes mellitus by bioinformatics analysis
- PMID: 31524257
- PMCID: PMC6691242
- DOI: 10.3892/mmr.2019.10522
Identification of core genes and pathways in type 2 diabetes mellitus by bioinformatics analysis
Abstract
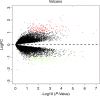
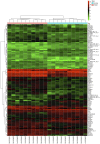
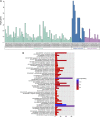
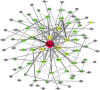
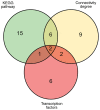
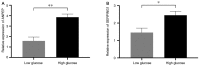
Type 2 diabetes mellitus (T2DM) is a metabolic disorder. Numerous proteins have been identified that are associated with the occurrence and development of T2DM. This study aimed to identify potential core genes and pathways involved in T2DM, through exhaustive bioinformatic analyses using GSE20966 microarray profiles of pancreatic β‑cells obtained from healthy controls and patients with T2DM. The original microarray data were downloaded from the Gene Expression Omnibus database. Data were processed by the limma package in R software and the differentially expressed genes (DEGs) were identified. Gene Ontology functional analysis and Kyoto Encyclopedia of Genes and Genomes pathway analysis were carried out to identify potential biological functions and pathways of the DEGs. Key transcription factors were identified using the WEB‑based GEne SeT AnaLysis Toolkit (WebGestalt) and Enrichr. The Search Tool for the Retrieval of Interacting Genes (STRING) database was used to establish a protein‑protein interaction (PPI) network for the DEGs. In total, 329 DEGs were involved in T2DM, with 208 upregulated genes enriched in pancreatic secretion and the complement and coagulation cascades, and 121 downregulated genes enriched in insulin secretion, carbohydrate digestion and absorption, and the Toll‑like receptor pathway. Furthermore, hepatocyte nuclear factor 1‑alpha (HNF1A), signal transducer and activator of transcription 3 (STAT3) and glucocorticoid receptor (GR) were key transcription factors in T2DM. Twenty important nodes were detected in the PPI network. Finally, two core genes, serpin family G member 1 (SERPING1) and alanyl aminopeptidase, membrane (ANPEP), were shown to be associated with the development of T2DM. On the whole, the findings of this study enhance our understanding of the potential molecular mechanisms of T2DM and provide potential targets for further research.
Figures


References
-
- Zhao Q, Zhang A, Zong W, An N, Zhang H, Luan Y, Sun H, Wang X, Cao H. Exploring potential biomarkers and determining the metabolic mechanism of type 2 diabetes mellitus using liquid chromatography coupled to high-resolution mass spectrometry. RSC Adv. 2017;7:44186. doi: 10.1039/C7RA05722A. - DOI
-
- Hoshino A, Ariyoshi M, Okawa Y, Kaimoto S, Uchihashi M, Fukai K, Iwai-Kanai E, Ikeda K, Ueyama T, Ogata T, Matoba S. Inhibition of p53 preserves Parkin-mediated mitophagy and pancreatic β-cell function in diabetes. Proc Natl Acad Sci USA. 2014;111:3116–3121. doi: 10.1073/pnas.1318951111. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

