Dynamic turnover of centromeres drives karyotype evolution in Drosophila
- PMID: 31524597
- PMCID: PMC6795482
- DOI: 10.7554/eLife.49002
Dynamic turnover of centromeres drives karyotype evolution in Drosophila
Abstract
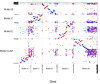
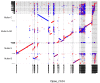
Centromeres are the basic unit for chromosome inheritance, but their evolutionary dynamics is poorly understood. We generate high-quality reference genomes for multiple Drosophila obscura group species to reconstruct karyotype evolution. All chromosomes in this lineage were ancestrally telocentric and the creation of metacentric chromosomes in some species was driven by de novo seeding of new centromeres at ancestrally gene-rich regions, independently of chromosomal rearrangements. The emergence of centromeres resulted in a drastic size increase due to repeat accumulation, and dozens of genes previously located in euchromatin are now embedded in pericentromeric heterochromatin. Metacentric chromosomes secondarily became telocentric in the pseudoobscura subgroup through centromere repositioning and a pericentric inversion. The former (peri)centric sequences left behind shrunk dramatically in size after their inactivation, yet contain remnants of their evolutionary past, including increased repeat-content and heterochromatic environment. Centromere movements are accompanied by rapid turnover of the major satellite DNA detected in (peri)centromeric regions.
Keywords: Drosophila obscura; centromere repositioning; chromosomes; evolutionary biology; gene expression; karyotype evolution.
© 2019, Bracewell et al.
Conflict of interest statement
RB, KC, MN, DB No competing interests declared
Figures



















References
-
- Ashburner M. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press: Cold Spring Harbor; 1989.
-
- Bachmann L, Raab M, Sperlich D. Satellite DNA and speciation: a species specific satellite DNA of Drosophila guanchel. Journal of Zoological Systematics and Evolutionary Research. 2009;27:84–93. doi: 10.1111/j.1439-0469.1989.tb00333.x. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

