Clonal ST131- H 22 Escherichia coli strains from a healthy pig and a human urinary tract infection carry highly similar resistance and virulence plasmids
- PMID: 31526455
- PMCID: PMC6807379
- DOI: 10.1099/mgen.0.000295
Clonal ST131- H 22 Escherichia coli strains from a healthy pig and a human urinary tract infection carry highly similar resistance and virulence plasmids
Abstract
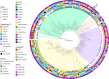
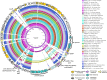
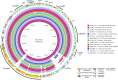
The interplay between food production animals, humans and the environment with respect to the transmission of drug-resistant pathogens is widely debated and poorly understood. Pandemic uropathogenic Escherichia coli ST131-H30Rx, with conserved fluoroquinolone and cephalosporin resistance, are not frequently identified in animals. However, the phylogenetic precursor lineage ST131-H22 in animals and associated meat products is being reported with increasing frequency. Here we characterized two highly related ST131-H22 strains, one from a healthy pig and the other from a human infection (in 2007 and 2009, respectively). We used both long and short genome sequencing and compared them to ST131-H22 genome sequences available in public repositories. Even within the context of H22 strains, the two strains in question were highly related, separated by only 20 core SNPs. Furthermore, they were closely related to a faecal strain isolated in 2010 from a geographically distinct, healthy human in New South Wales, Australia. The porcine and hospital strains carried highly similar HI2-ST3 multidrug resistant plasmids with differences in the hospital strain arising due to IS-mediated insertions and rearrangements. Near identical ColV plasmids were also present in both strains, further supporting their shared evolutionary history. This work highlights the importance of adopting a One Health approach to genomic surveillance to gain insights into pathogen evolution and spread.
Keywords: ColV; ExPEC; H22; HI2; ST131; fimH22; porcine commensal E. coli.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


Similar articles
-
Escherichia coli ST131-H22 as a Foodborne Uropathogen.mBio. 2018 Aug 28;9(4):e00470-18. doi: 10.1128/mBio.00470-18. mBio. 2018. PMID: 30154256 Free PMC article.
-
The ST131 Escherichia coli H22 subclone from human intestinal microbiota: Comparison of genomic and phenotypic traits with those of the globally successful H30 subclone.BMC Microbiol. 2017 Mar 27;17(1):71. doi: 10.1186/s12866-017-0984-8. BMC Microbiol. 2017. PMID: 28347271 Free PMC article.
-
Insights into a multidrug resistant Escherichia coli pathogen of the globally disseminated ST131 lineage: genome analysis and virulence mechanisms.PLoS One. 2011;6(10):e26578. doi: 10.1371/journal.pone.0026578. Epub 2011 Oct 28. PLoS One. 2011. PMID: 22053197 Free PMC article.
-
Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain.J Antimicrob Chemother. 2011 Jan;66(1):1-14. doi: 10.1093/jac/dkq415. Epub 2010 Nov 16. J Antimicrob Chemother. 2011. PMID: 21081548 Review.
-
Multidrug-resistant extraintestinal pathogenic Escherichia coli of sequence type ST131 in animals and foods.Vet Microbiol. 2011 Nov 21;153(1-2):99-108. doi: 10.1016/j.vetmic.2011.05.007. Epub 2011 May 13. Vet Microbiol. 2011. PMID: 21658865 Review.
Cited by
-
Genomic Characterisation of a Multiple Drug Resistant IncHI2 ST4 Plasmid in Escherichia coli ST744 in Australia.Microorganisms. 2020 Jun 14;8(6):896. doi: 10.3390/microorganisms8060896. Microorganisms. 2020. PMID: 32545892 Free PMC article.
-
Genomic comparisons of Escherichia coli ST131 from Australia.Microb Genom. 2021 Dec;7(12):000721. doi: 10.1099/mgen.0.000721. Microb Genom. 2021. PMID: 34910614 Free PMC article.
-
Genomic analysis of plasmid content in food isolates of E. coli strongly supports its role as a reservoir for the horizontal transfer of virulence and antibiotic resistance genes.Plasmid. 2022 Sep-Nov;123-124:102650. doi: 10.1016/j.plasmid.2022.102650. Epub 2022 Sep 18. Plasmid. 2022. PMID: 36130651 Free PMC article.
-
Genomic Analysis of an I1 Plasmid Hosting a sul3-Class 1 Integron and blaSHV-12 within an Unusual Escherichia coli ST297 from Urban Wildlife.Microorganisms. 2022 Jul 10;10(7):1387. doi: 10.3390/microorganisms10071387. Microorganisms. 2022. PMID: 35889108 Free PMC article.
-
Identification of genes influencing the evolution of Escherichia coli ST372 in dogs and humans.Microb Genom. 2023 Feb;9(2):mgen000930. doi: 10.1099/mgen.0.000930. Microb Genom. 2023. PMID: 36752777 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

