Pseudomonas aeruginosa Ethanol Oxidation by AdhA in Low-Oxygen Environments
- PMID: 31527114
- PMCID: PMC6832066
- DOI: 10.1128/JB.00393-19
Pseudomonas aeruginosa Ethanol Oxidation by AdhA in Low-Oxygen Environments
Abstract
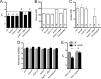
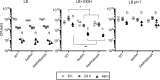
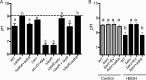
Pseudomonas aeruginosa has a broad metabolic repertoire that facilitates its coexistence with different microbes. Many microbes secrete products that P. aeruginosa can then catabolize, including ethanol, a common fermentation product. Here, we show that under oxygen-limiting conditions P. aeruginosa utilizes AdhA, an NAD-linked alcohol dehydrogenase, as a previously undescribed means for ethanol catabolism. In a rich medium containing ethanol, AdhA, but not the previously described PQQ-linked alcohol dehydrogenase, ExaA, oxidizes ethanol and leads to the accumulation of acetate in culture supernatants. AdhA-dependent acetate accumulation and the accompanying decrease in pH promote P. aeruginosa survival in LB-grown stationary-phase cultures. The transcription of adhA is elevated by hypoxia and under anoxic conditions, and we show that it is regulated by the Anr transcription factor. We have shown that lasR mutants, which lack an important quorum sensing regulator, have higher levels of Anr-regulated transcripts under low-oxygen conditions than their wild-type counterparts. Here, we show that a lasR mutant, when grown with ethanol, has an even larger decrease in pH than the wild type (WT) that is dependent on both anr and adhA The large increase in AdhA activity is similar to that of a strain expressing a hyperactive Anr-D149A variant. Ethanol catabolism in P. aeruginosa by AdhA supports growth on ethanol as a sole carbon source and electron donor in oxygen-limited settings and in cells growing by denitrification under anoxic conditions. This is the first demonstration of a physiological role for AdhA in ethanol oxidation in P. aeruginosaIMPORTANCE Ethanol is a common product of microbial fermentation, and the Pseudomonas aeruginosa response to and utilization of ethanol are relevant to our understanding of its role in microbial communities. Here, we report that the putative alcohol dehydrogenase AdhA is responsible for ethanol catabolism and acetate accumulation under low-oxygen conditions and that it is regulated by Anr.
Keywords: AdhA; ExaA; Pseudomonas aeruginosa; ethanol; lasR.
Copyright © 2019 American Society for Microbiology.
Figures
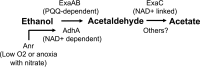
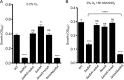
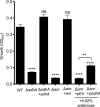
Similar articles
-
Pseudomonas aeruginosa lasR mutant fitness in microoxia is supported by an Anr-regulated oxygen-binding hemerythrin.Proc Natl Acad Sci U S A. 2020 Feb 11;117(6):3167-3173. doi: 10.1073/pnas.1917576117. Epub 2020 Jan 24. Proc Natl Acad Sci U S A. 2020. PMID: 31980538 Free PMC article.
-
Links between Anr and Quorum Sensing in Pseudomonas aeruginosa Biofilms.J Bacteriol. 2015 Sep;197(17):2810-20. doi: 10.1128/JB.00182-15. Epub 2015 Jun 15. J Bacteriol. 2015. PMID: 26078448 Free PMC article.
-
Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa.J Bacteriol. 2000 Dec;182(24):6940-9. doi: 10.1128/JB.182.24.6940-6949.2000. J Bacteriol. 2000. PMID: 11092854 Free PMC article.
-
The alcohol dehydrogenase gene adhA in Corynebacterium glutamicum is subject to carbon catabolite repression.J Bacteriol. 2007 Oct;189(20):7408-16. doi: 10.1128/JB.00791-07. Epub 2007 Aug 10. J Bacteriol. 2007. PMID: 17693518 Free PMC article.
-
The ethanol oxidation system and its regulation in Pseudomonas aeruginosa.Biochim Biophys Acta. 2003 Apr 11;1647(1-2):98-102. doi: 10.1016/s1570-9639(03)00066-9. Biochim Biophys Acta. 2003. PMID: 12686116 Review.
Cited by
-
Pseudomonas aeruginosa lasR mutant fitness in microoxia is supported by an Anr-regulated oxygen-binding hemerythrin.Proc Natl Acad Sci U S A. 2020 Feb 11;117(6):3167-3173. doi: 10.1073/pnas.1917576117. Epub 2020 Jan 24. Proc Natl Acad Sci U S A. 2020. PMID: 31980538 Free PMC article.
-
Global transcriptional response of Pseudomonas aeruginosa to UVA radiation.Photochem Photobiol Sci. 2024 Nov;23(11):2029-2044. doi: 10.1007/s43630-024-00649-9. Epub 2024 Oct 29. Photochem Photobiol Sci. 2024. PMID: 39470974
-
Metabolic basis for the evolution of a common pathogenic Pseudomonas aeruginosa variant.Elife. 2022 May 3;11:e76555. doi: 10.7554/eLife.76555. Elife. 2022. PMID: 35502894 Free PMC article.
-
Cometabolism of Ethanol in Azospirillum brasilense Sp7 Is Mediated by Fructose and Glycerol and Regulated Negatively by an Alternative Sigma Factor RpoH2.J Bacteriol. 2021 Nov 19;203(24):e0026921. doi: 10.1128/JB.00269-21. Epub 2021 Sep 27. J Bacteriol. 2021. PMID: 34570625 Free PMC article.
-
Mechanisms controlling bacterial infection in myeloid cells under hypoxic conditions.Cell Mol Life Sci. 2021 Mar;78(5):1887-1907. doi: 10.1007/s00018-020-03684-8. Epub 2020 Oct 30. Cell Mol Life Sci. 2021. PMID: 33125509 Free PMC article. Review.
References
-
- Montuschi P, Paris D, Melck D, Lucidi V, Ciabattoni G, Raia V, Calabrese C, Bush A, Barnes PJ, Motta A. 2012. NMR spectroscopy metabolomic profiling of exhaled breath condensate in patients with stable and unstable cystic fibrosis. Thorax 67:222–228. doi:10.1136/thoraxjnl-2011-200072. - DOI - PubMed
-
- Chen AI, Dolben EF, Okegbe C, Harty CE, Golub Y, Thao S, Ha DG, Willger SD, O'Toole GA, Harwood CS, Dietrich LEP, Hogan DA. 2014. Candida albicans ethanol stimulates Pseudomonas aeruginosa WspR-controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog 10:e1004480. doi:10.1371/journal.ppat.1004480. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

