Centrosomal cohesion deficits as cellular biomarker in lymphoblastoid cell lines from LRRK2 Parkinson's disease patients
- PMID: 31527116
- PMCID: PMC6792036
- DOI: 10.1042/BCJ20190315
Centrosomal cohesion deficits as cellular biomarker in lymphoblastoid cell lines from LRRK2 Parkinson's disease patients
Abstract
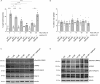
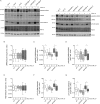
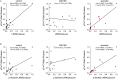
Leucine-rich repeat kinase 2 (LRRK2) is a promising therapeutic target for the treatment of Parkinson's disease (PD), and orally bioavailable, brain penetrant and highly potent LRRK2 kinase inhibitors are in early stages of clinical testing. Detection of LRRK2 phosphorylation, as well as phosphorylation of Rab10, a LRRK2 kinase substrate, have been proposed as target engagement biomarkers for LRRK2 inhibitor clinical trials. However, these readouts do not seem able to stratify patients based on enhanced LRRK2 kinase activity. Here, we describe a robust cell biological assay based on centrosomal cohesion alterations which were observed in peripheral blood mononuclear cell-derived lymphoblastoid cell lines (LCLs) from patients with G2019S LRRK2 mutations as compared with healthy controls, and could also be detected in a subset of sporadic PD patient samples. We suggest that LCLs may be a valuable resource for LRRK2 research, and that determination of centrosomal cohesion deficits may assist in the stratification of a subset of sporadic PD patients.
Keywords: Rab protein; centrosome cohesion; leucine-rich repeat kinase; protein phosphorylation.
© 2019 The Author(s).
Conflict of interest statement
The Authors declare that there are no competing interests associated with the manuscript.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

