Heme binding to human CLOCK affects interactions with the E-box
- PMID: 31527239
- PMCID: PMC6778266
- DOI: 10.1073/pnas.1905216116
Heme binding to human CLOCK affects interactions with the E-box
Abstract
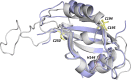
The circadian clock is an endogenous time-keeping system that is ubiquitous in animals and plants as well as some bacteria. In mammals, the clock regulates the sleep-wake cycle via 2 basic helix-loop-helix PER-ARNT-SIM (bHLH-PAS) domain proteins-CLOCK and BMAL1. There is emerging evidence to suggest that heme affects circadian control, through binding of heme to various circadian proteins, but the mechanisms of regulation are largely unknown. In this work we examine the interaction of heme with human CLOCK (hCLOCK). We present a crystal structure for the PAS-A domain of hCLOCK, and we examine heme binding to the PAS-A and PAS-B domains. UV-visible and electron paramagnetic resonance spectroscopies are consistent with a bis-histidine ligated heme species in solution in the oxidized (ferric) PAS-A protein, and by mutagenesis we identify His144 as a ligand to the heme. There is evidence for flexibility in the heme pocket, which may give rise to an additional Cys axial ligand at 20K (His/Cys coordination). Using DNA binding assays, we demonstrate that heme disrupts binding of CLOCK to its E-box DNA target. Evidence is presented for a conformationally mobile protein framework, which is linked to changes in heme ligation and which has the capacity to affect binding to the E-box. Within the hCLOCK structural framework, this would provide a mechanism for heme-dependent transcriptional regulation.
Keywords: CLOCK; circadian; heme.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Intermolecular recognition revealed by the complex structure of human CLOCK-BMAL1 basic helix-loop-helix domains with E-box DNA.Cell Res. 2013 Feb;23(2):213-24. doi: 10.1038/cr.2012.170. Epub 2012 Dec 11. Cell Res. 2013. PMID: 23229515 Free PMC article.
-
Formation of a repressive complex in the mammalian circadian clock is mediated by the secondary pocket of CRY1.Proc Natl Acad Sci U S A. 2017 Feb 14;114(7):1560-1565. doi: 10.1073/pnas.1615310114. Epub 2017 Jan 31. Proc Natl Acad Sci U S A. 2017. PMID: 28143926 Free PMC article.
-
Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex.Science. 2012 Jul 13;337(6091):189-94. doi: 10.1126/science.1222804. Epub 2012 May 31. Science. 2012. PMID: 22653727 Free PMC article.
-
PAS Dimerization at the Nexus of the Mammalian Circadian Clock.J Mol Biol. 2024 Feb 1;436(3):168341. doi: 10.1016/j.jmb.2023.168341. Epub 2023 Nov 2. J Mol Biol. 2024. PMID: 37924861 Free PMC article. Review.
-
Structural characterization of mammalian bHLH-PAS transcription factors.Curr Opin Struct Biol. 2017 Apr;43:1-9. doi: 10.1016/j.sbi.2016.09.011. Epub 2016 Oct 6. Curr Opin Struct Biol. 2017. PMID: 27721191 Free PMC article. Review.
Cited by
-
Multi-Modal Regulation of Circadian Physiology by Interactive Features of Biological Clocks.Biology (Basel). 2021 Dec 24;11(1):21. doi: 10.3390/biology11010021. Biology (Basel). 2021. PMID: 35053019 Free PMC article. Review.
-
Understanding the Logistics for the Distribution of Heme in Cells.JACS Au. 2021 Aug 10;1(10):1541-1555. doi: 10.1021/jacsau.1c00288. eCollection 2021 Oct 25. JACS Au. 2021. PMID: 34723258 Free PMC article. Review.
-
Endogenous Hemoprotein-Dependent Signaling Pathways of Nitric Oxide and Nitrite.Inorg Chem. 2021 Nov 1;60(21):15918-15940. doi: 10.1021/acs.inorgchem.1c01048. Epub 2021 Jul 27. Inorg Chem. 2021. PMID: 34313417 Free PMC article. Review.
-
Circadian Genes Expression Patterns in Disorders Due to Enzyme Deficiencies in the Heme Biosynthetic Pathway.Biomedicines. 2022 Dec 9;10(12):3198. doi: 10.3390/biomedicines10123198. Biomedicines. 2022. PMID: 36551954 Free PMC article.
-
Role of TET1-mediated epigenetic modulation in Alzheimer's disease.Neurobiol Dis. 2023 Sep;185:106257. doi: 10.1016/j.nbd.2023.106257. Epub 2023 Aug 8. Neurobiol Dis. 2023. PMID: 37562656 Free PMC article.
References
-
- Gallego M., Virshup D. M., Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 8, 139–148 (2007). - PubMed
-
- Shimizu T., et al. , Gaseous O2, NO, and CO in signal transduction: Structure and function relationships of heme-based gas sensors and heme-redox sensors. Chem. Rev. 115, 6491–6533 (2015). - PubMed
-
- Verma A., Hirsch D. J., Glatt C. E., Ronnett G. V., Snyder S. H., Carbon monoxide: A putative neural messenger. Science 259, 381–384 (1993). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

