Organization of fast and slow chromatin revealed by single-nucleosome dynamics
- PMID: 31527274
- PMCID: PMC6778247
- DOI: 10.1073/pnas.1907342116
Organization of fast and slow chromatin revealed by single-nucleosome dynamics
Abstract
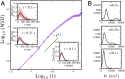
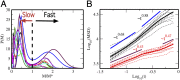
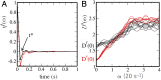
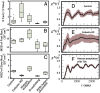
Understanding chromatin organization and dynamics is important, since they crucially affect DNA functions. In this study, we investigate chromatin dynamics by statistically analyzing single-nucleosome movement in living human cells. Bimodal nature of the mean square displacement distribution of nucleosomes allows for a natural categorization of the nucleosomes as fast and slow. Analyses of the nucleosome-nucleosome correlation functions within these categories along with the density of vibrational modes show that the nucleosomes form dynamically correlated fluid regions (i.e., dynamic domains of fast and slow nucleosomes). Perturbed nucleosome dynamics by global histone acetylation or cohesin inactivation indicate that nucleosome-nucleosome interactions along with tethering of chromatin chains organize nucleosomes into fast and slow dynamic domains. A simple polymer model is introduced, which shows the consistency of this dynamic domain picture. Statistical analyses of single-nucleosome movement provide rich information on how chromatin is dynamically organized in a fluid manner in living cells.
Keywords: chromatin domains; cohesin; histone; live cell imaging; nucleosomes.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Bickmore W. A., The spatial organization of the human genome. Annu. Rev. Genom. Hum. Genet. 14, 67–84 (2013). - PubMed
-
- Cardoso M. C., Schneider K., Martin R. M., Leonhardt H., Structure, function and dynamics of nuclear subcompartments. Curr. Opin. Cell Biol. 24, 79–85 (2012). - PubMed
-
- Maeshima K., Ide S., Hibino K., Sasai M., Liquid-like behavior of chromatin. Curr. Opin. Genet. Dev. 37, 36–45 (2016). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

