The role of sugar-backbone heterogeneity and chimeras in the simultaneous emergence of RNA and DNA
- PMID: 31527850
- PMCID: PMC6815252
- DOI: 10.1038/s41557-019-0322-x
The role of sugar-backbone heterogeneity and chimeras in the simultaneous emergence of RNA and DNA
Abstract
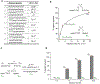
Hypotheses of the origins of RNA and DNA are generally centred on the prebiotic synthesis of a pristine system (pre-RNA or RNA), which gives rise to its descendent. However, a lack of specificity in the synthesis of genetic polymers would probably result in chimeric sequences; the roles and fate of such sequences are unknown. Here, we show that chimeras, exemplified by mixed threose nucleic acid (TNA)-RNA and RNA-DNA oligonucleotides, preferentially bind to, and act as templates for, homogeneous TNA, RNA and DNA ligands. The chimeric templates can act as a catalyst that mediates the ligation of oligomers to give homogeneous backbone sequences, and the regeneration of the chimeric templates potentiates a scenario for a possible cross-catalytic cycle with amplification. This process provides a proof-of-principle demonstration of a heterogeneity-to-homogeneity scenario and also gives credence to the idea that DNA could appear concurrently with RNA, instead of being its later descendent.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
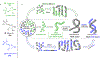




References
-
- Gilbert W Origin of Life - the RNA World. Nature 319, 618–618 (1986).
-
- Joyce GF & Orgel LE Progress toward understanding the origin of the RNA world. Cold Spring Harbor Monograph Series 43, 23–56 (2006).
-
- Gesteland RF, Cech TR & Atkins JF The RNA world: the nature of modern RNA suggests a prebiotic RNA. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor,1999).
-
- Schwartz A Origins of the RNA world In Brack A (Ed.), The Molecular Origins of Life: Assembling Pieces of the Puzzle. pp. 237–254 (Cambridge: Cambridge University Press, 1998).
-
- Orgel LE Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol 39, 99–123 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

