Antibiotic-resistant Escherichia coli and Salmonella spp. associated with dairy cattle and farm environment having public health significance
- PMID: 31528022
- PMCID: PMC6702575
- DOI: 10.14202/vetworld.2019.984-993
Antibiotic-resistant Escherichia coli and Salmonella spp. associated with dairy cattle and farm environment having public health significance
Abstract
Aim: The present study was carried out to determine load of total bacteria, Escherichia coli and Salmonella spp. in dairy farm and its environmental components. In addition, the antibiogram profile of the isolated bacteria having public health impact was also determined along with identification of virulence and resistance genes by polymerase chain reaction (PCR) under a one-health approach.
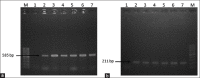
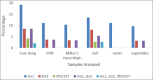
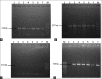
Materials and methods: A total of 240 samples of six types (cow dung - 15, milk - 10, milkers' hand wash - 10, soil - 10 water - 5, and vegetables - 10) were collected from four dairy farms. For enumeration, the samples were cultured onto plate count agar, eosin methylene blue, and xylose-lysine deoxycholate agar and the isolation and identification of the E. coli and Salmonella spp. were performed based on morphology, cultural, staining, and biochemical properties followed by PCR.The pathogenic strains of E. coli stx1, stx2, and rfbO157 were also identified through PCR. The isolates were subjected to antimicrobial susceptibility test against 12 commonly used antibiotics by disk diffusion method. Detection of antibiotic resistance genes ereA, tetA, tetB, and SHV were performed by PCR.
Results: The mean total bacterial count, E. coli and Salmonella spp. count in the samples ranged from 4.54±0.05 to 8.65±0.06, 3.62±0.07 to 7.04±0.48, and 2.52±0.08 to 5.87±0.05 log colony-forming unit/g or ml, respectively. Out of 240 samples, 180 (75%) isolates of E. coli and 136 (56.67%) isolates of Salmonella spp. were recovered through cultural and molecular tests. Among the 180 E. coli isolates, 47 (26.11%) were found positive for the presence of all the three virulent genes, of which stx1 was the most prevalent (13.33%). Only three isolates were identified as enterohemorrhagic E. coli. Antibiotic sensitivity test revealed that both E. coli and Salmonella spp. were found highly resistant to azithromycin, tetracycline, erythromycin, oxytetracycline, and ertapenem and susceptible to gentamycin, ciprofloxacin, and imipenem. Among the four antibiotic resistance genes, the most observable was tetA (80.51-84.74%) in E. coli and Salmonella spp. and SHV genes were the lowest one (22.06-25%).
Conclusion: Dairy farm and their environmental components carry antibiotic-resistant pathogenic E. coli and Salmonella spp. that are potential threat for human health which requires a one-health approach to combat the threat.
Keywords: Escherichia coli; Salmonella spp; antibiotic resistance genes; carbapenem resistance; dairy farm; one-health; virulence.
Figures
Similar articles
-
Antibiotic Resistance Profiles of Escherichia coli and Salmonella spp. Isolated From Dairy Farms and Surroundings in a Rural Area of Western Anatolia, Turkey.Cureus. 2024 Aug 2;16(8):e65996. doi: 10.7759/cureus.65996. eCollection 2024 Aug. Cureus. 2024. PMID: 39221349 Free PMC article.
-
Isolation of multidrug-resistant Escherichia coli and Salmonella spp. from sulfonamide-treated diarrheic calves.Vet World. 2022 Dec;15(12):2870-2876. doi: 10.14202/vetworld.2022.2870-2876. Epub 2022 Dec 16. Vet World. 2022. PMID: 36718340 Free PMC article.
-
Multidrug-resistant Escherichia coli and Salmonella spp. isolated from pigeons.Vet World. 2020 Oct;13(10):2156-2165. doi: 10.14202/vetworld.2020.2156-2165. Epub 2020 Oct 15. Vet World. 2020. PMID: 33281350 Free PMC article.
-
Salmonella spp. in Domestic Ruminants, Evaluation of Antimicrobial Resistance Based on the One Health Approach-A Systematic Review and Meta-Analysis.Vet Sci. 2024 Jul 14;11(7):315. doi: 10.3390/vetsci11070315. Vet Sci. 2024. PMID: 39057999 Free PMC article. Review.
-
Microbial Contamination and Antibiotic Resistance in Fresh Produce and Agro-Ecosystems in South Asia-A Systematic Review.Microorganisms. 2024 Nov 8;12(11):2267. doi: 10.3390/microorganisms12112267. Microorganisms. 2024. PMID: 39597656 Free PMC article. Review.
Cited by
-
High Occurrence of Multidrug-Resistant Escherichia coli Strains in Bovine Fecal Samples from Healthy Cows Serves as Rich Reservoir for AMR Transmission.Antibiotics (Basel). 2022 Dec 26;12(1):37. doi: 10.3390/antibiotics12010037. Antibiotics (Basel). 2022. PMID: 36671238 Free PMC article.
-
Prevalence and antibiotic resistance of Salmonella spp. in South Punjab-Pakistan.PLoS One. 2020 Nov 19;15(11):e0232382. doi: 10.1371/journal.pone.0232382. eCollection 2020. PLoS One. 2020. Retraction in: PLoS One. 2022 Oct 21;17(10):e0275948. doi: 10.1371/journal.pone.0275948. PMID: 33211713 Free PMC article. Retracted.
-
Epidemiology and Molecular Characterisation of Multidrug-Resistant Escherichia coli Isolated from Cow Milk.Vet Sci. 2024 Nov 29;11(12):609. doi: 10.3390/vetsci11120609. Vet Sci. 2024. PMID: 39728949 Free PMC article.
-
Isolation, Identification and Antimicrobial Resistance Profile of Salmonella in Raw cow milk & its products in Bishoftu city, central Ethiopia: implication for public health.One Health Outlook. 2025 Mar 15;7(1):10. doi: 10.1186/s42522-025-00134-y. One Health Outlook. 2025. PMID: 40087724 Free PMC article.
-
Epidemiology and molecular characterisation of multidrug-resistant Escherichia coli isolated from chicken meat.PLoS One. 2025 May 14;20(5):e0323909. doi: 10.1371/journal.pone.0323909. eCollection 2025. PLoS One. 2025. PMID: 40367070 Free PMC article.
References
-
- Tule A, Hassani U. Colonization with antibiotic-resistant E. coli in commensal fecal flora of newborns. Int. J. Curr. Microbiol. Appl. Sci. 2017;6(5):1623–1629.
-
- Mckenna M. The Coming Cost of Superbugs:10 Million Deaths Per Year. 2014. [Last accesed on 10.01.2019]. Available from: http://www.wired.com/2014/12/oneill-rptamr .
LinkOut - more resources
Full Text Sources
Miscellaneous
