IDH2 reprograms mitochondrial dynamics in cancer through a HIF-1α - regulated pseudohypoxic state
- PMID: 31530011
- PMCID: PMC6894043
- DOI: 10.1096/fj.201901366R
IDH2 reprograms mitochondrial dynamics in cancer through a HIF-1α - regulated pseudohypoxic state
Abstract
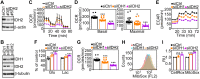
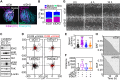
The role of mitochondria in cancer continues to be debated and paradoxically implicated in opposing functions in tumor growth and tumor suppression. To understand this dichotomy, we explored the function of mitochondrial isocitrate dehydrogenase (IDH)2, a tricarboxylic acid cycle enzyme mutated in subsets of acute leukemias and gliomas, in cancer. Silencing of IDH2 in prostate cancer cells impaired oxidative bioenergetics, elevated reactive oxygen species (ROS) production, and promoted exaggerated mitochondrial dynamics. This was associated with increased subcellular mitochondrial trafficking, turnover of membrane focal adhesion complexes, and enhanced tumor cell migration and invasion, without changes in cell cycle progression. Mechanistically, loss of IDH2 caused ROS-dependent stabilization of hypoxia-inducible factor-1α in normoxia, which was required for increased mitochondrial trafficking and tumor cell movements. Therefore, IDH2 is a dual regulator of cancer bioenergetics and tumor cell motility. This pathway may reprogram mitochondrial dynamics to differentially adjust energy production or promote tumor cell invasion in response to microenvironment conditions.-Wang, Y., Agarwal, E., Bertolini, I., Ghosh, J. C., Seo, J. H., Altieri, D. C. IDH2 reprograms mitochondrial dynamics in cancer through a HIF-1α-regulated pseudohypoxic state.
Keywords: HIF-1α metastasis; ROS; mitochondria; tumor cell motility.
Conflict of interest statement
The authors thank James Hayden and Frederick Keeney (The Wistar Institute Imaging Core Facility) for assistance with time-lapse videomicroscopy. This work was supported by U.S. National Institutes of Health, National Cancer Institute Grants P01 CA140043 and R35 CA220446 (to D.C.A.). The support for Shared Resources utilized in this study was provided by Cancer Center Support Grant (CCSG) P30 CA010815. The authors declare no conflicts of interest.
Figures





References
-
- Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 - PubMed
-
- Gatenby R. A., Gillies R. J. (2004) Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4, 891–899 - PubMed
-
- Lehuédé C., Dupuy F., Rabinovitch R., Jones R. G., Siegel P. M. (2016) Metabolic plasticity as a determinant of tumor growth and metastasis. Cancer Res. 76, 5201–5208 - PubMed
-
- Tabassum D. P., Polyak K. (2015) Tumorigenesis: it takes a village. Nat. Rev. Cancer 15, 473–483 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

