Prospecting for microbial α- N-acetylgalactosaminidases yields a new class of GH31 O-glycanase
- PMID: 31530641
- PMCID: PMC6827296
- DOI: 10.1074/jbc.RA119.010628
Prospecting for microbial α- N-acetylgalactosaminidases yields a new class of GH31 O-glycanase
Abstract
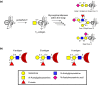
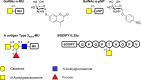
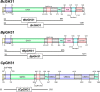
α-Linked GalNAc (α-GalNAc) is most notably found at the nonreducing terminus of the blood type-determining A-antigen and as the initial point of attachment to the peptide backbone in mucin-type O-glycans. However, despite their ubiquity in saccharolytic microbe-rich environments such as the human gut, relatively few α-N-acetylgalactosaminidases are known. Here, to discover and characterize novel microbial enzymes that hydrolyze α-GalNAc, we screened small-insert libraries containing metagenomic DNA from the human gut microbiome. Using a simple fluorogenic glycoside substrate, we identified and characterized a glycoside hydrolase 109 (GH109) that is active on blood type A-antigen, along with a new subfamily of glycoside hydrolase 31 (GH31) that specifically cleaves the initial α-GalNAc from mucin-type O-glycans. This represents a new activity in this GH family and a potentially useful new enzyme class for analysis or modification of O-glycans on protein or cell surfaces.
Keywords: N-acetylgalactosamine (alpha-GalNAc); O-glycanase; O-glycoprotein; blood; glycobiology; glycopeptide cleavage; glycoprotein; glycosidase; glycoside hydrolase 109 (GH109); glycoside hydrolase 31 (GH31); human gut microbiome; metagenomic analysis; metagenomics; mucin.
© 2019 Rahfeld et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures
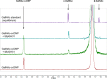
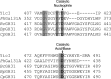
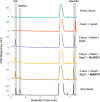



References
-
- Skovierová H, Larrouy-Maumus G., Pham H., Belanová M., Barilone N., Dasgupta A., Mikusová K., Gicquel B., Gilleron M., Brennan P. J., Puzo G., Nigou J., and Jackson M. (2010) Biosynthetic origin of the galactosamine substituent of arabinogalactan in Mycobacterium tuberculosis. J. Biol. Chem. 285, 41348–41355 10.1074/jbc.M110.188110 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

