A Multi-Layered Study on Harmonic Oscillations in Mammalian Genomics and Proteomics
- PMID: 31533246
- PMCID: PMC6770795
- DOI: 10.3390/ijms20184585
A Multi-Layered Study on Harmonic Oscillations in Mammalian Genomics and Proteomics
Abstract
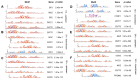
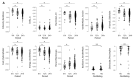
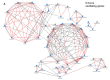
Cellular, organ, and whole animal physiology show temporal variation predominantly featuring 24-h (circadian) periodicity. Time-course mRNA gene expression profiling in mouse liver showed two subsets of genes oscillating at the second (12-h) and third (8-h) harmonic of the prime (24-h) frequency. The aim of our study was to identify specific genomic, proteomic, and functional properties of ultradian and circadian subsets. We found hallmarks of the three oscillating gene subsets, including different (i) functional annotation, (ii) proteomic and electrochemical features, and (iii) transcription factor binding motifs in upstream regions of 8-h and 12-h oscillating genes that seemingly allow the link of the ultradian gene sets to a known circadian network. Our multifaceted bioinformatics analysis of circadian and ultradian genes suggests that the different rhythmicity of gene expression impacts physiological outcomes and may be related to transcriptional, translational and post-translational dynamics, as well as to phylogenetic and evolutionary components.
Keywords: biological clock; circadian rhythms; electrochemical features; rhythmic gene expression; rhythmic protein expression; ultradian rhythms.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Systematic Analysis of Mouse Genome Reveals Distinct Evolutionary and Functional Properties Among Circadian and Ultradian Genes.Front Physiol. 2018 Aug 23;9:1178. doi: 10.3389/fphys.2018.01178. eCollection 2018. Front Physiol. 2018. PMID: 30190679 Free PMC article.
-
Mining for novel candidate clock genes in the circadian regulatory network.BMC Syst Biol. 2015 Nov 14;9:78. doi: 10.1186/s12918-015-0227-2. BMC Syst Biol. 2015. PMID: 26576534 Free PMC article.
-
Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames.Genome Res. 2015 Dec;25(12):1848-59. doi: 10.1101/gr.195404.115. Epub 2015 Oct 20. Genome Res. 2015. PMID: 26486724 Free PMC article.
-
Genetics and molecular biology of rhythms in Drosophila and other insects.Adv Genet. 2003;48:1-280. doi: 10.1016/s0065-2660(03)48000-0. Adv Genet. 2003. PMID: 12593455 Review.
-
Functional genomics and proteomics in the clinical neurosciences: data mining and bioinformatics.Prog Brain Res. 2006;158:83-108. doi: 10.1016/S0079-6123(06)58004-5. Prog Brain Res. 2006. PMID: 17027692 Review.
Cited by
-
Molecular World Today and Tomorrow: Recent Trends in Biological Sciences.Int J Mol Sci. 2024 Mar 6;25(5):3068. doi: 10.3390/ijms25053068. Int J Mol Sci. 2024. PMID: 38474313 Free PMC article.
-
Chronobiology Meets Quantum Biology: A New Paradigm Overlooking the Horizon?Front Physiol. 2022 Jul 6;13:892582. doi: 10.3389/fphys.2022.892582. eCollection 2022. Front Physiol. 2022. PMID: 35874510 Free PMC article. Review.
-
Preservation of ∼12-h ultradian rhythms of gene expression of mRNA and protein metabolism in the absence of canonical circadian clock.Front Physiol. 2023 May 30;14:1195001. doi: 10.3389/fphys.2023.1195001. eCollection 2023. Front Physiol. 2023. PMID: 37324401 Free PMC article.
-
An Optimal Time for Treatment-Predicting Circadian Time by Machine Learning and Mathematical Modelling.Cancers (Basel). 2020 Oct 23;12(11):3103. doi: 10.3390/cancers12113103. Cancers (Basel). 2020. PMID: 33114254 Free PMC article. Review.
-
Preservation of ∼12-hour ultradian rhythms of gene expression of mRNA and protein metabolism in the absence of canonical circadian clock.bioRxiv [Preprint]. 2023 May 2:2023.05.01.538977. doi: 10.1101/2023.05.01.538977. bioRxiv. 2023. Update in: Front Physiol. 2023 May 30;14:1195001. doi: 10.3389/fphys.2023.1195001. PMID: 37205336 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

