Metaviromics Reveals Unknown Viral Diversity in the Biting Midge Culicoides impunctatus
- PMID: 31533247
- PMCID: PMC6784199
- DOI: 10.3390/v11090865
Metaviromics Reveals Unknown Viral Diversity in the Biting Midge Culicoides impunctatus
Abstract
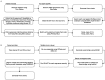
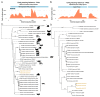
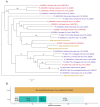
Biting midges (Culicoides species) are vectors of arboviruses and were responsible for the emergence and spread of Schmallenberg virus (SBV) in Europe in 2011 and are likely to be involved in the emergence of other arboviruses in Europe. Improved surveillance and better understanding of risks require a better understanding of the circulating viral diversity in these biting insects. In this study, we expand the sequence space of RNA viruses by identifying a number of novel RNA viruses from Culicoides impunctatus (biting midge) using a meta-transcriptomic approach. A novel metaviromic pipeline called MetaViC was developed specifically to identify novel virus sequence signatures from high throughput sequencing (HTS) datasets in the absence of a known host genome. MetaViC is a protein centric pipeline that looks for specific protein signatures in the reads and contigs generated as part of the pipeline. Several novel viruses, including an alphanodavirus with both segments, a novel relative of the Hubei sobemo-like virus 49, two rhabdo-like viruses and a chuvirus, were identified in the Scottish midge samples. The newly identified viruses were found to be phylogenetically distinct to those previous known. These findings expand our current knowledge of viral diversity in arthropods and especially in these understudied disease vectors.
Keywords: Culicoides impunctatus; RNA viruses; metaviromics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Investigation of RNA Viruses in Culicoides Latreille, 1809 (Diptera: Ceratopogonidae) in a Mining Complex in the Southeastern Region of the Brazilian Amazon.Viruses. 2024 Nov 29;16(12):1862. doi: 10.3390/v16121862. Viruses. 2024. PMID: 39772171 Free PMC article.
-
Profiling of RNA Viruses in Biting Midges (Ceratopogonidae) and Related Diptera from Kenya Using Metagenomics and Metabarcoding Analysis.mSphere. 2021 Oct 27;6(5):e0055121. doi: 10.1128/mSphere.00551-21. Epub 2021 Oct 13. mSphere. 2021. PMID: 34643419 Free PMC article.
-
Characterisation of the virome of Culicoides brevitarsis Kieffer (Diptera: Ceratopogonidae), a vector of bluetongue virus in Australia.J Gen Virol. 2025 Feb;106(2):002076. doi: 10.1099/jgv.0.002076. J Gen Virol. 2025. PMID: 39976626 Free PMC article.
-
Culicoides Biting Midges-Underestimated Vectors for Arboviruses of Public Health and Veterinary Importance.Viruses. 2019 Apr 24;11(4):376. doi: 10.3390/v11040376. Viruses. 2019. PMID: 31022868 Free PMC article. Review.
-
Culicoides biting midges, arboviruses and public health in Europe.Antiviral Res. 2013 Oct;100(1):102-13. doi: 10.1016/j.antiviral.2013.07.020. Epub 2013 Aug 8. Antiviral Res. 2013. PMID: 23933421 Review.
Cited by
-
Jingchuvirales: a New Taxonomical Framework for a Rapidly Expanding Order of Unusual Monjiviricete Viruses Broadly Distributed among Arthropod Subphyla.Appl Environ Microbiol. 2022 Mar 22;88(6):e0195421. doi: 10.1128/AEM.01954-21. Epub 2022 Feb 2. Appl Environ Microbiol. 2022. PMID: 35108077 Free PMC article. Review.
-
Metavirome Analysis and Identification of Midge-Borne Viruses from Yunnan Province, China, in 2021.Viruses. 2023 Aug 26;15(9):1817. doi: 10.3390/v15091817. Viruses. 2023. PMID: 37766224 Free PMC article.
-
Meta-Viromic Sequencing Reveals Virome Characteristics of Mosquitoes and Culicoides on Zhoushan Island, China.Microbiol Spectr. 2023 Feb 14;11(1):e0268822. doi: 10.1128/spectrum.02688-22. Epub 2023 Jan 18. Microbiol Spectr. 2023. PMID: 36651764 Free PMC article.
-
Exploiting insect-specific viruses as a novel strategy to control vector-borne disease.Curr Opin Insect Sci. 2020 Jun;39:50-56. doi: 10.1016/j.cois.2020.02.005. Epub 2020 Feb 28. Curr Opin Insect Sci. 2020. PMID: 32278312 Free PMC article. Review.
-
The Role of Hematophagous Arthropods, Other than Mosquitoes and Ticks, in Arbovirus Transmission.Viruses. 2025 Jun 30;17(7):932. doi: 10.3390/v17070932. Viruses. 2025. PMID: 40733550 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

