A novel protein descriptor for the prediction of drug binding sites
- PMID: 31533611
- PMCID: PMC6749706
- DOI: 10.1186/s12859-019-3058-0
A novel protein descriptor for the prediction of drug binding sites
Abstract
Background: Binding sites are the pockets of proteins that can bind drugs; the discovery of these pockets is a critical step in drug design. With the help of computers, protein pockets prediction can save manpower and financial resources.
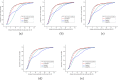
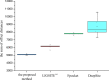
Results: In this paper, a novel protein descriptor for the prediction of binding sites is proposed. Information on non-bonded interactions in the three-dimensional structure of a protein is captured by a combination of geometry-based and energy-based methods. Moreover, due to the rapid development of deep learning, all binding features are extracted to generate three-dimensional grids that are fed into a convolution neural network. Two datasets were introduced into the experiment. The sc-PDB dataset was used for descriptor extraction and binding site prediction, and the PDBbind dataset was used only for testing and verification of the generalization of the method. The comparison with previous methods shows that the proposed descriptor is effective in predicting the binding sites.
Conclusions: A new protein descriptor is proposed for the prediction of the drug binding sites of proteins. This method combines the three-dimensional structure of a protein and non-bonded interactions with small molecules to involve important factors influencing the formation of binding site. Analysis of the experiments indicates that the descriptor is robust for site prediction.
Keywords: Binding sites prediction; Deep learning; Molecule descriptor; Protein pockets.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures









References
-
- Lu Pinyi, Bevan David R., Leber Andrew, Hontecillas Raquel, Tubau-Juni Nuria, Bassaganya-Riera Josep. Accelerated Path to Cures. Cham: Springer International Publishing; 2018. Computer-Aided Drug Discovery; pp. 7–24.
-
- Liu Z, Li Y, Han L, Li J, Liu J, Zhao Z, Nie W, Liu Y, Wang R. Pdb-wide collection of binding data: current status of the PDBbind database. Bioinformatics. 2014;31(3):405–12. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

