Hepatitis C Virus Infection Is Inhibited by a Noncanonical Antiviral Signaling Pathway Targeted by NS3-NS4A
- PMID: 31534039
- PMCID: PMC6854490
- DOI: 10.1128/JVI.00725-19
Hepatitis C Virus Infection Is Inhibited by a Noncanonical Antiviral Signaling Pathway Targeted by NS3-NS4A
Abstract
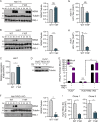
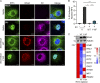
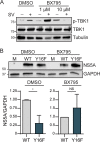
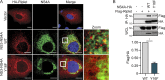
The hepatitis C virus (HCV) NS3-NS4A protease complex is required for viral replication and is the major viral innate immune evasion factor. NS3-NS4A evades antiviral innate immunity by inactivating several proteins, including MAVS, the signaling adaptor for RIG-I and MDA5, and Riplet, an E3 ubiquitin ligase that activates RIG-I. Here, we identified a Tyr-16-Phe (Y16F) change in the NS4A transmembrane domain that prevents NS3-NS4A targeting of Riplet but not MAVS. This Y16F substitution reduces HCV replication in Huh7 cells, but not in Huh-7.5 cells, known to lack RIG-I signaling. Surprisingly, deletion of RIG-I in Huh7 cells did not restore Y16F viral replication. Rather, we found that Huh-7.5 cells lack Riplet expression and that the addition of Riplet to these cells reduced HCV Y16F replication, whereas the addition of Riplet lacking the RING domain restored HCV Y16F replication. In addition, TBK1 inhibition or IRF3 deletion in Huh7 cells was sufficient to restore HCV Y16F replication, and the Y16F protease lacked the ability to prevent IRF3 activation or interferon induction. Taken together, these data reveal that the NS4A Y16 residue regulates a noncanonical Riplet-TBK1-IRF3-dependent, but RIG-I-MAVS-independent, signaling pathway that limits HCV infection.IMPORTANCE The HCV NS3-NS4A protease complex facilitates viral replication by cleaving and inactivating the antiviral innate immune signaling proteins MAVS and Riplet, which are essential for RIG-I activation. NS3-NS4A therefore prevents IRF3 activation and interferon induction during HCV infection. Here, we uncover an amino acid residue within the NS4A transmembrane domain that is essential for inactivation of Riplet but does not affect MAVS cleavage by NS3-NS4A. Our study reveals that Riplet is involved in a RIG-I- and MAVS-independent signaling pathway that activates IRF3 and that this pathway is normally inactivated by NS3-NS4A during HCV infection. Our study selectively uncouples these distinct regulatory mechanisms within NS3-NS4A and defines a new role for Riplet in the antiviral response to HCV. Since Riplet is known to be inhibited by other RNA viruses, such as such influenza A virus, this innate immune signaling pathway may also be important in controlling other RNA virus infections.
Keywords: HCV; MAVS; NS3-NS4A; Riplet; antiviral innate immunity; immune evasion; protease.
Copyright © 2019 American Society for Microbiology.
Figures




Similar articles
-
Control of innate immune signaling and membrane targeting by the Hepatitis C virus NS3/4A protease are governed by the NS3 helix α0.J Virol. 2012 Mar;86(6):3112-20. doi: 10.1128/JVI.06727-11. Epub 2012 Jan 11. J Virol. 2012. PMID: 22238314 Free PMC article.
-
Restoration of the activated Rig-I pathway in hepatitis C virus (HCV) replicon cells by HCV protease, polymerase, and NS5A inhibitors in vitro at clinically relevant concentrations.Antimicrob Agents Chemother. 2013 Sep;57(9):4417-26. doi: 10.1128/AAC.00399-13. Epub 2013 Jul 8. Antimicrob Agents Chemother. 2013. PMID: 23836176 Free PMC article.
-
Dengue Virus NS Proteins Inhibit RIG-I/MAVS Signaling by Blocking TBK1/IRF3 Phosphorylation: Dengue Virus Serotype 1 NS4A Is a Unique Interferon-Regulating Virulence Determinant.mBio. 2015 May 12;6(3):e00553-15. doi: 10.1128/mBio.00553-15. mBio. 2015. PMID: 25968648 Free PMC article.
-
[Chronic hepatitis C virus infection attenuates host antiviral innate immune response].Nihon Rinsho. 2015 Feb;73(2):234-8. Nihon Rinsho. 2015. PMID: 25764676 Review. Japanese.
-
Hepatitis A and hepatitis C viruses: divergent infection outcomes marked by similarities in induction and evasion of interferon responses.Semin Liver Dis. 2010 Nov;30(4):319-32. doi: 10.1055/s-0030-1267534. Epub 2010 Oct 19. Semin Liver Dis. 2010. PMID: 20960373 Review.
Cited by
-
Host Innate Immunity Against Hepatitis Viruses and Viral Immune Evasion.Front Microbiol. 2021 Nov 3;12:740464. doi: 10.3389/fmicb.2021.740464. eCollection 2021. Front Microbiol. 2021. PMID: 34803956 Free PMC article. Review.
-
Synthetic RIG-I-Agonist RNA Induces Death of Hepatocellular Carcinoma Cells.J Interferon Cytokine Res. 2025 Apr;45(4):119-132. doi: 10.1089/jir.2024.0195. Epub 2025 Feb 13. J Interferon Cytokine Res. 2025. PMID: 39945619
-
Flaviviridae Nonstructural Proteins: The Role in Molecular Mechanisms of Triggering Inflammation.Viruses. 2022 Aug 18;14(8):1808. doi: 10.3390/v14081808. Viruses. 2022. PMID: 36016430 Free PMC article. Review.
-
Hepatitis C Virus: Evading the Intracellular Innate Immunity.J Clin Med. 2020 Mar 13;9(3):790. doi: 10.3390/jcm9030790. J Clin Med. 2020. PMID: 32183176 Free PMC article. Review.
-
Reconstitution of interferon regulatory factor 7 expression restores interferon beta induction in Huh7 cells.J Virol. 2025 Jun 17;99(6):e0070325. doi: 10.1128/jvi.00703-25. Epub 2025 May 23. J Virol. 2025. PMID: 40407345 Free PMC article.
References
-
- World Health Organization. 2017. Global hepatitis report. World Health Organization, Geneva, Switzerland: License: CC BY-NC-SA 3.0 IGO.
-
- Bartenschlager R, Baumert TF, Bukh J, Houghton M, Lemon SM, Lindenbach BD, Lohmann V, Moradpour D, Pietschmann T, Rice CM, Thimme R, Wakita T. 2018. Critical challenges and emerging opportunities in hepatitis C virus research in an era of potent antiviral therapy: considerations for scientists and funding agencies. Virus Res 248:53–62. doi:10.1016/j.virusres.2018.02.016. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

