α-Ketoglutarate links p53 to cell fate during tumour suppression
- PMID: 31534224
- PMCID: PMC6830448
- DOI: 10.1038/s41586-019-1577-5
α-Ketoglutarate links p53 to cell fate during tumour suppression
Abstract
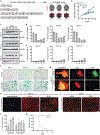
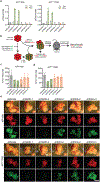
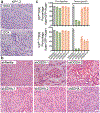
The tumour suppressor TP53 is mutated in the majority of human cancers, and in over 70% of pancreatic ductal adenocarcinoma (PDAC)1,2. Wild-type p53 accumulates in response to cellular stress, and regulates gene expression to alter cell fate and prevent tumour development2. Wild-type p53 is also known to modulate cellular metabolic pathways3, although p53-dependent metabolic alterations that constrain cancer progression remain poorly understood. Here we find that p53 remodels cancer-cell metabolism to enforce changes in chromatin and gene expression that favour a premalignant cell fate. Restoring p53 function in cancer cells derived from KRAS-mutant mouse models of PDAC leads to the accumulation of α-ketoglutarate (αKG, also known as 2-oxoglutarate), a metabolite that also serves as an obligate substrate for a subset of chromatin-modifying enzymes. p53 induces transcriptional programs that are characteristic of premalignant differentiation, and this effect can be partially recapitulated by the addition of cell-permeable αKG. Increased levels of the αKG-dependent chromatin modification 5-hydroxymethylcytosine (5hmC) accompany the tumour-cell differentiation that is triggered by p53, whereas decreased 5hmC characterizes the transition from premalignant to de-differentiated malignant lesions that is associated with mutations in Trp53. Enforcing the accumulation of αKG in p53-deficient PDAC cells through the inhibition of oxoglutarate dehydrogenase-an enzyme of the tricarboxylic acid cycle-specifically results in increased 5hmC, tumour-cell differentiation and decreased tumour-cell fitness. Conversely, increasing the intracellular levels of succinate (a competitive inhibitor of αKG-dependent dioxygenases) blunts p53-driven tumour suppression. These data suggest that αKG is an effector of p53-mediated tumour suppression, and that the accumulation of αKG in p53-deficient tumours can drive tumour-cell differentiation and antagonize malignant progression.
Conflict of interest statement
Figures



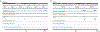







Comment in
-
Suppressed fate.Nat Rev Cancer. 2019 Nov;19(11):610. doi: 10.1038/s41568-019-0215-8. Nat Rev Cancer. 2019. PMID: 31578518 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

