A robust human norovirus replication model in zebrafish larvae
- PMID: 31536612
- PMCID: PMC6752765
- DOI: 10.1371/journal.ppat.1008009
A robust human norovirus replication model in zebrafish larvae
Abstract
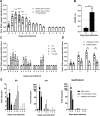
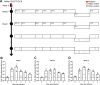
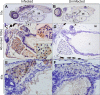
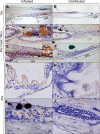
Human noroviruses (HuNoVs) are the most common cause of foodborne illness, with a societal cost of $60 billion and 219,000 deaths/year. The lack of robust small animal models has significantly hindered the understanding of norovirus biology and the development of effective therapeutics. Here we report that HuNoV GI and GII replicate to high titers in zebrafish (Danio rerio) larvae; replication peaks at day 2 post infection and is detectable for at least 6 days. The virus (HuNoV GII.4) could be passaged from larva to larva two consecutive times. HuNoV is detected in cells of the hematopoietic lineage and the intestine, supporting the notion of a dual tropism. Antiviral treatment reduces HuNoV replication by >2 log10, showing that this model is suited for antiviral studies. Zebrafish larvae constitute a simple and robust replication model that will largely facilitate studies of HuNoV biology and the development of antiviral strategies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Hemming M, Rasanen S, Huhti L, Paloniemi M, Salminen M, Vesikari T. Major reduction of rotavirus, but not norovirus, gastroenteritis in children seen in hospital after the introduction of RotaTeq vaccine into the National Immunization Programme in Finland. European journal of pediatrics. 2013;172(6):739–46. Epub 2013/01/31. 10.1007/s00431-013-1945-3 . - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

