Label-Free Assay of Protein Tyrosine Phosphatase Activity in Single Cells
- PMID: 31536703
- PMCID: PMC6889211
- DOI: 10.1021/acs.analchem.9b03640
Label-Free Assay of Protein Tyrosine Phosphatase Activity in Single Cells
Abstract
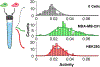
Populations of cells exhibit variations in biochemical activity, resulting from many factors including random stochastic variability in protein production, metabolic and cell-cycle states, regulatory mechanisms, and external signaling. The development of methods for the analysis of single cells has allowed for the measurement and understanding of this inherent heterogeneity, yet methods for measuring protein activities on the single-cell scale lag behind their genetic analysis counterparts and typically report on expression rather than activity. This paper presents an approach to measure protein tyrosine phosphatase (PTP) activity in individual cells using self-assembled monolayers for matrix-assisted laser desorption/ionization mass spectrometry. Using flow cytometry, individual cells are first sorted into a well plate containing lysis buffer and a phosphopeptide substrate. After lysis and incubation-during which the PTP enzymes act on the peptide substrate-the reaction substrate and product are immobilized onto arrays of self-assembled monolayers, which are then analyzed using mass spectrometry. PTP activities from thousands of individual cells were measured and their distributions analyzed. This work demonstrates a general method for measuring enzyme activities in lysates derived from individual cells and will contribute to the understanding of cellular heterogeneity in a variety of contexts.
Figures
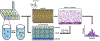


Similar articles
-
Profiling Protein Tyrosine Phosphatase Specificity with Self-Assembled Monolayers for Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry and Peptide Arrays.ACS Comb Sci. 2019 Nov 11;21(11):760-769. doi: 10.1021/acscombsci.9b00152. Epub 2019 Oct 24. ACS Comb Sci. 2019. PMID: 31553163 Free PMC article.
-
Combining SAMDI Mass Spectrometry and Peptide Arrays to Profile Phosphatase Activities.Methods Enzymol. 2018;607:389-403. doi: 10.1016/bs.mie.2018.04.021. Epub 2018 Jun 19. Methods Enzymol. 2018. PMID: 30149867 Free PMC article.
-
Assay of protein tyrosine phosphatases by using matrix-assisted laser desorption ionization time-of-flight mass spectrometry.Anal Biochem. 2001 May 1;292(1):51-8. doi: 10.1006/abio.2001.5071. Anal Biochem. 2001. PMID: 11319817
-
Cellular biochemistry methods for investigating protein tyrosine phosphatases.Antioxid Redox Signal. 2014 May 10;20(14):2160-78. doi: 10.1089/ars.2013.5731. Epub 2014 Feb 25. Antioxid Redox Signal. 2014. PMID: 24294920 Free PMC article. Review.
-
Pseudophosphatases: methods of analysis and physiological functions.Methods. 2014 Jan 15;65(2):207-18. doi: 10.1016/j.ymeth.2013.09.009. Epub 2013 Sep 21. Methods. 2014. PMID: 24064037 Review.
Cited by
-
Analysis of Phosphatase Activity in a Droplet-Based Microfluidic Chip.Biosensors (Basel). 2022 Sep 8;12(9):740. doi: 10.3390/bios12090740. Biosensors (Basel). 2022. PMID: 36140125 Free PMC article.
-
Narratives of Undergraduate Research, Mentorship, and Teaching at UCLA.Pure Appl Chem. 2021;93(2):207-221. doi: 10.1515/pac-2020-1007. Epub 2021 Feb 8. Pure Appl Chem. 2021. PMID: 33935303 Free PMC article.
-
A fluorescent probe for monitoring PTP-PEST enzymatic activity.Analyst. 2020 Oct 21;145(20):6713-6718. doi: 10.1039/d0an00993h. Epub 2020 Aug 19. Analyst. 2020. PMID: 32812952 Free PMC article.
-
Exploring the Ligand Preferences of the PHD1 Domain of Histone Demethylase KDM5A Reveals Tolerance for Modifications of the Q5 Residue of Histone 3.ACS Chem Biol. 2021 Jan 15;16(1):205-213. doi: 10.1021/acschembio.0c00891. Epub 2020 Dec 14. ACS Chem Biol. 2021. PMID: 33314922 Free PMC article.
-
A high-throughput SAMDI-mass spectrometry assay for isocitrate dehydrogenase 1.Analyst. 2020 Jun 7;145(11):3899-3908. doi: 10.1039/d0an00174k. Epub 2020 Apr 16. Analyst. 2020. PMID: 32297889 Free PMC article.
References
-
- Zhang L; Vertes A Single-cell mass spectrometry approaches to explore cellular heterogeneity. Angew. Chem. Int. Ed 2018, 57, 4466–4477. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

