Extending Tests of Hardy-Weinberg Equilibrium to Structured Populations
- PMID: 31537622
- PMCID: PMC6827367
- DOI: 10.1534/genetics.119.302370
Extending Tests of Hardy-Weinberg Equilibrium to Structured Populations
Abstract
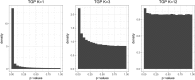
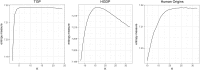
Testing for Hardy-Weinberg equilibrium (HWE) is an important component in almost all analyses of population genetic data. Genetic markers that violate HWE are often treated as special cases; for example, they may be flagged as possible genotyping errors, or they may be investigated more closely for evolutionary signatures of interest. The presence of population structure is one reason why genetic markers may fail a test of HWE. This is problematic because almost all natural populations studied in the modern setting show some degree of structure. Therefore, it is important to be able to detect deviations from HWE for reasons other than structure. To this end, we extend statistical tests of HWE to allow for population structure, which we call a test of "structural HWE." Additionally, our new test allows one to automatically choose tuning parameters and identify accurate models of structure. We demonstrate our approach on several important studies, provide theoretical justification for the test, and present empirical evidence for its utility. We anticipate the proposed test will be useful in a broad range of analyses of genome-wide population genetic data.
Keywords: Chi-square test; HWE; admixed; admixture; population genetics; quality control; random mating; significance test.
Copyright © 2019 Hao and Storey.
Figures




References
-
- Billingsley P., 2012. Probability and Measure, Ed. 4 Wiley, Hoboken, NJ.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

