A kindlin-3-leupaxin-paxillin signaling pathway regulates podosome stability
- PMID: 31537712
- PMCID: PMC6781449
- DOI: 10.1083/jcb.201903109
A kindlin-3-leupaxin-paxillin signaling pathway regulates podosome stability
Abstract
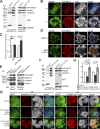
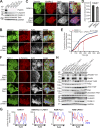
Binding of kindlins to integrins is required for integrin activation, stable ligand binding, and subsequent intracellular signaling. How hematopoietic kindlin-3 contributes to the assembly and stability of the adhesion complex is not known. Here we report that kindlin-3 recruits leupaxin into podosomes and thereby regulates paxillin phosphorylation and podosome turnover. We demonstrate that the activity of the protein tyrosine phosphatase PTP-PEST, which controls paxillin phosphorylation, requires leupaxin. In contrast, despite sharing the same binding mode with leupaxin, paxillin recruitment into podosomes is kindlin-3 independent. Instead, we found paxillin together with talin and vinculin in initial adhesion patches of kindlin-3-null cells. Surprisingly, despite its presence in these early adhesion patches, podosomes can form in the absence of paxillin or any paxillin member. In conclusion, our findings show that kindlin-3 not only activates and clusters integrins into podosomes but also regulates their lifetime by recruiting leupaxin, which controls PTP-PEST activity and thereby paxillin phosphorylation and downstream signaling.
© 2019 Klapproth et al.
Figures
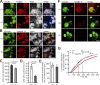
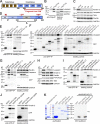
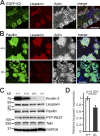
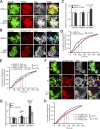




Comment in
-
Kindlin-3 stokes the life span of podosomes.J Cell Biol. 2019 Oct 7;218(10):3166-3167. doi: 10.1083/jcb.201909096. Epub 2019 Sep 23. J Cell Biol. 2019. PMID: 31548283 Free PMC article.
Similar articles
-
Kindlin-3 stokes the life span of podosomes.J Cell Biol. 2019 Oct 7;218(10):3166-3167. doi: 10.1083/jcb.201909096. Epub 2019 Sep 23. J Cell Biol. 2019. PMID: 31548283 Free PMC article.
-
LIM domain-containing adaptor, leupaxin, localizes in focal adhesion and suppresses the integrin-induced tyrosine phosphorylation of paxillin.Cancer Sci. 2010 Feb;101(2):363-8. doi: 10.1111/j.1349-7006.2009.01398.x. Epub 2009 Oct 12. Cancer Sci. 2010. PMID: 19917054 Free PMC article.
-
Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin.Elife. 2016 Jan 27;5:e10130. doi: 10.7554/eLife.10130. Elife. 2016. PMID: 26821125 Free PMC article.
-
Paxillin and Kindlin: Research Progress and Biological Functions.Biomolecules. 2025 Jan 24;15(2):173. doi: 10.3390/biom15020173. Biomolecules. 2025. PMID: 40001476 Free PMC article. Review.
-
Paxillin and its role in the aging process of skin cells.Postepy Hig Med Dosw (Online). 2016 Oct 6;70(0):1087-1094. doi: 10.5604/17322693.1221385. Postepy Hig Med Dosw (Online). 2016. PMID: 27708212 Review.
Cited by
-
Conformationally active integrin endocytosis and traffic: why, where, when and how?Biochem Soc Trans. 2020 Feb 28;48(1):83-93. doi: 10.1042/BST20190309. Biochem Soc Trans. 2020. PMID: 32065228 Free PMC article. Review.
-
Kindlin-3 stokes the life span of podosomes.J Cell Biol. 2019 Oct 7;218(10):3166-3167. doi: 10.1083/jcb.201909096. Epub 2019 Sep 23. J Cell Biol. 2019. PMID: 31548283 Free PMC article.
-
Site-specific phosphorylation regulates the functions of kindlin-3 in a variety of cells.Life Sci Alliance. 2020 Feb 5;3(3):e201900594. doi: 10.26508/lsa.201900594. Print 2019 Mar. Life Sci Alliance. 2020. PMID: 32024667 Free PMC article.
-
A flexible loop in the paxillin LIM3 domain mediates its direct binding to integrin β subunits.PLoS Biol. 2024 Sep 4;22(9):e3002757. doi: 10.1371/journal.pbio.3002757. eCollection 2024 Sep. PLoS Biol. 2024. PMID: 39231388 Free PMC article.
-
Decreased Tiam1-mediated Rac1 activation is responsible for impaired directional persistence of chondrocyte migration in microtia.J Cell Mol Med. 2024 Jun;28(11):e18443. doi: 10.1111/jcmm.18443. J Cell Mol Med. 2024. PMID: 38837873 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

