An African Salmonella Typhimurium ST313 sublineage with extensive drug-resistance and signatures of host adaptation
- PMID: 31537784
- PMCID: PMC6753159
- DOI: 10.1038/s41467-019-11844-z
An African Salmonella Typhimurium ST313 sublineage with extensive drug-resistance and signatures of host adaptation
Abstract
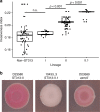
Bloodstream infections by Salmonella enterica serovar Typhimurium constitute a major health burden in sub-Saharan Africa (SSA). These invasive non-typhoidal (iNTS) infections are dominated by isolates of the antibiotic resistance-associated sequence type (ST) 313. Here, we report emergence of ST313 sublineage II.1 in the Democratic Republic of the Congo. Sublineage II.1 exhibits extensive drug resistance, involving a combination of multidrug resistance, extended spectrum β-lactamase production and azithromycin resistance. ST313 lineage II.1 isolates harbour an IncHI2 plasmid we name pSTm-ST313-II.1, with one isolate also exhibiting decreased ciprofloxacin susceptibility. Whole genome sequencing reveals that ST313 II.1 isolates have accumulated genetic signatures potentially associated with altered pathogenicity and host adaptation, related to changes observed in biofilm formation and metabolic capacity. Sublineage II.1 emerged at the beginning of the 21st century and is involved in on-going outbreaks. Our data provide evidence of further evolution within the ST313 clade associated with iNTS in SSA.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

