NPC1 regulates ER contacts with endocytic organelles to mediate cholesterol egress
- PMID: 31537798
- PMCID: PMC6753064
- DOI: 10.1038/s41467-019-12152-2
NPC1 regulates ER contacts with endocytic organelles to mediate cholesterol egress
Abstract
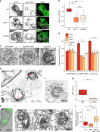
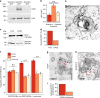
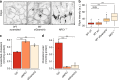
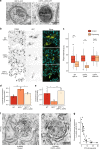
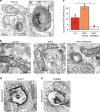
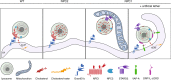
Transport of dietary cholesterol from endocytic organelles to the endoplasmic reticulum (ER) is essential for cholesterol homoeostasis, but the mechanism and regulation of this transport remains poorly defined. Membrane contact sites (MCS), microdomains of close membrane apposition, are gaining attention as important platforms for non-vesicular, inter-organellar communication. Here we investigate the impact of ER-endocytic organelle MCS on cholesterol transport. We report a role for Niemann-Pick type C protein 1 (NPC1) in tethering ER-endocytic organelle MCS where it interacts with the ER-localised sterol transport protein Gramd1b to regulate cholesterol egress. We show that artificially tethering MCS rescues the cholesterol accumulation that characterises NPC1-deficient cells, consistent with direct lysosome to ER cholesterol transport across MCS. Finally, we identify an expanded population of lysosome-mitochondria MCS in cells depleted of NPC1 or Gramd1b that is dependent on the late endosomal sterol-binding protein STARD3, likely underlying the mitochondrial cholesterol accumulation in NPC1-deficient cells.
Conflict of interest statement
F.P. is consultant to Actelion, Orphazyme and IntraBio and co-founder of IntraBio. Other authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

