Genomic Organization of the B3-Domain Transcription Factor Family in Grapevine (Vitis vinifera L.) and Expression during Seed Development in Seedless and Seeded Cultivars
- PMID: 31540007
- PMCID: PMC6770561
- DOI: 10.3390/ijms20184553
Genomic Organization of the B3-Domain Transcription Factor Family in Grapevine (Vitis vinifera L.) and Expression during Seed Development in Seedless and Seeded Cultivars
Abstract
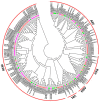
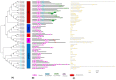
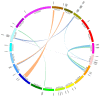
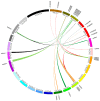
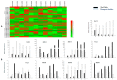
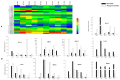
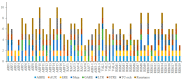
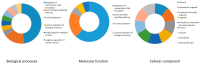
Members of the plant-specific B3-domain transcription factor family have important and varied functions, especially with respect to vegetative and reproductive growth. Although B3 genes have been studied in many other plants, there is limited information on the genomic organization and expression of B3 genes in grapevine (Vitis vinifera L.). In this study, we identified 50 B3 genes in the grapevine genome and analyzed these genes in terms of chromosomal location and syntenic relationships, intron-exon organization, and promoter cis-element content. We also analyzed the presumed proteins in terms of domain structure and phylogenetic relationships. Based on the results, we classified these genes into five subfamilies. The syntenic relationships suggest that approximately half of the genes resulted from genome duplication, contributing to the expansion of the B3 family in grapevine. The analysis of cis-element composition suggested that most of these genes may function in response to hormones, light, and stress. We also analyzed expression of members of the B3 family in various structures of grapevine plants, including the seed during seed development. Many B3 genes were expressed preferentially in one or more structures of the developed plant, suggesting specific roles in growth and development. Furthermore, several of the genes were expressed differentially in early developing seeds from representative seeded and seedless cultivars, suggesting a role in seed development or abortion. The results of this study provide a foundation for functional analysis of B3 genes and new resources for future molecular breeding of grapevine.
Keywords: B3 superfamily; Vitis vinifera; expression analysis; ovule abortion; transcription factor.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Genome-Wide Characterization and Expression Profiling of GASA Genes during Different Stages of Seed Development in Grapevine (Vitis vinifera L.) Predict Their Involvement in Seed Development.Int J Mol Sci. 2020 Feb 6;21(3):1088. doi: 10.3390/ijms21031088. Int J Mol Sci. 2020. PMID: 32041336 Free PMC article.
-
Genome-wide identification and expression analysis of the B-box transcription factor gene family in grapevine (Vitis vinifera L.).BMC Genomics. 2021 Mar 29;22(1):221. doi: 10.1186/s12864-021-07479-4. BMC Genomics. 2021. PMID: 33781207 Free PMC article.
-
Genome-wide identification and expression analyses of the homeobox transcription factor family during ovule development in seedless and seeded grapes.Sci Rep. 2017 Oct 3;7(1):12638. doi: 10.1038/s41598-017-12988-y. Sci Rep. 2017. PMID: 28974771 Free PMC article.
-
Precision breeding of grapevine (Vitis vinifera L.) for improved traits.Plant Sci. 2014 Nov;228:3-10. doi: 10.1016/j.plantsci.2014.03.023. Epub 2014 Apr 6. Plant Sci. 2014. PMID: 25438781 Review.
-
Back to the Origins: Background and Perspectives of Grapevine Domestication.Int J Mol Sci. 2021 Apr 26;22(9):4518. doi: 10.3390/ijms22094518. Int J Mol Sci. 2021. PMID: 33926017 Free PMC article. Review.
Cited by
-
Genome-wide association studies dissect the genetic architecture of seed shape and size in common bean.G3 (Bethesda). 2022 Apr 4;12(4):jkac048. doi: 10.1093/g3journal/jkac048. G3 (Bethesda). 2022. PMID: 35218340 Free PMC article.
-
Genome-wide identification of the soybean cytokinin oxidase/dehydrogenase gene family and its diverse roles in response to multiple abiotic stress.Front Plant Sci. 2023 Apr 17;14:1163219. doi: 10.3389/fpls.2023.1163219. eCollection 2023. Front Plant Sci. 2023. PMID: 37139113 Free PMC article.
-
Exploration of the B3 transcription factor superfamily in Aquilaria sinensis reveal their involvement in seed recalcitrance and agarwood formation.PLoS One. 2023 Nov 16;18(11):e0294358. doi: 10.1371/journal.pone.0294358. eCollection 2023. PLoS One. 2023. PMID: 37972007 Free PMC article.
-
Genetic changes in the genus Vitis and the domestication of vine.Front Plant Sci. 2023 Feb 28;13:1019311. doi: 10.3389/fpls.2022.1019311. eCollection 2022. Front Plant Sci. 2023. PMID: 36926258 Free PMC article. Review.
-
Genome-Wide Characterization and Expression Profiling of GASA Genes during Different Stages of Seed Development in Grapevine (Vitis vinifera L.) Predict Their Involvement in Seed Development.Int J Mol Sci. 2020 Feb 6;21(3):1088. doi: 10.3390/ijms21031088. Int J Mol Sci. 2020. PMID: 32041336 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

