Development of a Fragment-Based Screening Assay for the Focal Adhesion Targeting Domain Using SPR and NMR
- PMID: 31540099
- PMCID: PMC6766811
- DOI: 10.3390/molecules24183352
Development of a Fragment-Based Screening Assay for the Focal Adhesion Targeting Domain Using SPR and NMR
Abstract
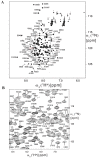
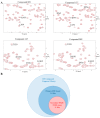
The Focal Adhesion Targeting (FAT) domain of Focal Adhesion Kinase (FAK) is a promising drug target since FAK is overexpressed in many malignancies and promotes cancer cell metastasis. The FAT domain serves as a scaffolding protein, and its interaction with the protein paxillin localizes FAK to focal adhesions. Various studies have highlighted the importance of FAT-paxillin binding in tumor growth, cell invasion, and metastasis. Targeting this interaction through high-throughput screening (HTS) provides a challenge due to the large and complex binding interface. In this report, we describe a novel approach to targeting FAT through fragment-based drug discovery (FBDD). We developed two fragment-based screening assays-a primary SPR assay and a secondary heteronuclear single quantum coherence nuclear magnetic resonance (HSQC-NMR) assay. For SPR, we designed an AviTag construct, optimized SPR buffer conditions, and created mutant controls. For NMR, resonance backbone assignments of the human FAT domain were obtained for the HSQC assay. A 189-compound fragment library from Enamine was screened through our primary SPR assay to demonstrate the feasibility of a FAT-FBDD pipeline, with 19 initial hit compounds. A final total of 11 validated hits were identified after secondary screening on NMR. This screening pipeline is the first FBDD screen of the FAT domain reported and represents a valid method for further drug discovery efforts on this difficult target.
Keywords: FAT domain; focal adhesion kinase; fragment-based drug discovery; nuclear magnetic resonance; surface plasmon resonance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Owens L.V., Xu L., Craven R.J., Dent G.A., Weiner T.M., Kornberg L., Liu E.T., Cance W.G. Overexpression of the focal adhesion kinase (p125fak) in invasive human tumors. Cancer Res. 1995;55:2752–2755. - PubMed
-
- Lark A.L., Livasy C.A., Calvo B., Caskey L., Moore D.T., Yang X., Cance W.G. Overexpression of focal adhesion kinase in primary colorectal carcinomas and colorectal liver metastases: Immunohistochemistry and real-time pcr analyses. Clin. Cancer Res. 2003;9:215–222. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

