Recombination in Enteroviruses, a Multi-Step Modular Evolutionary Process
- PMID: 31540135
- PMCID: PMC6784155
- DOI: 10.3390/v11090859
Recombination in Enteroviruses, a Multi-Step Modular Evolutionary Process
Abstract
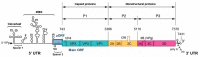
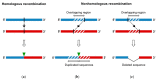
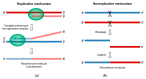
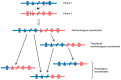
RNA recombination is a major driving force in the evolution and genetic architecture shaping of enteroviruses. In particular, intertypic recombination is implicated in the emergence of most pathogenic circulating vaccine-derived polioviruses, which have caused numerous outbreaks of paralytic poliomyelitis worldwide. Recent experimental studies that relied on recombination cellular systems mimicking natural genetic exchanges between enteroviruses provided new insights into the molecular mechanisms of enterovirus recombination and enabled to define a new model of genetic plasticity for enteroviruses. Homologous intertypic recombinant enteroviruses that were observed in nature would be the final products of a multi-step process, during which precursor nonhomologous recombinant genomes are generated through an initial inter-genomic RNA recombination event and can then evolve into a diversity of fitter homologous recombinant genomes over subsequent intra-genomic rearrangements. Moreover, these experimental studies demonstrated that the enterovirus genome could be defined as a combination of genomic modules that can be preferentially exchanged through recombination, and enabled defining the boundaries of these recombination modules. These results provided the first experimental evidence supporting the theoretical model of enterovirus modular evolution previously elaborated from phylogenetic studies of circulating enterovirus strains. This review summarizes our current knowledge regarding the mechanisms of recombination in enteroviruses and presents a new evolutionary process that may apply to other RNA viruses.
Keywords: RNA virus; emergence; enterovirus; recombination; viral evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Pallansch M., Roos R. Enteroviruses: Polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott Williams and Wilkins; Philadelphia, PA, USA: 2007. pp. 839–893.
-
- Yang T.-T., Huang L.-M., Lu C.-Y., Kao C.-L., Lee W.-T., Lee P.-I., Chen C.-M., Huang F.-Y., Lee C.-Y., Chang L.-Y. Clinical features and factors of unfavorable outcomes for non-polio enterovirus infection of the central nervous system in northern Taiwan, 1994–2003. J. Microbiol. Immunol. Infect. 2005;38:417–424. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

