Key Topics in Molecular Docking for Drug Design
- PMID: 31540192
- PMCID: PMC6769580
- DOI: 10.3390/ijms20184574
Key Topics in Molecular Docking for Drug Design
Abstract
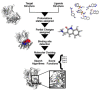
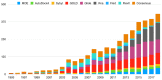
Molecular docking has been widely employed as a fast and inexpensive technique in the past decades, both in academic and industrial settings. Although this discipline has now had enough time to consolidate, many aspects remain challenging and there is still not a straightforward and accurate route to readily pinpoint true ligands among a set of molecules, nor to identify with precision the correct ligand conformation within the binding pocket of a given target molecule. Nevertheless, new approaches continue to be developed and the volume of published works grows at a rapid pace. In this review, we present an overview of the method and attempt to summarise recent developments regarding four main aspects of molecular docking approaches: (i) the available benchmarking sets, highlighting their advantages and caveats, (ii) the advances in consensus methods, (iii) recent algorithms and applications using fragment-based approaches, and (iv) the use of machine learning algorithms in molecular docking. These recent developments incrementally contribute to an increase in accuracy and are expected, given time, and together with advances in computing power and hardware capability, to eventually accomplish the full potential of this area.
Keywords: benchmarking sets; computer-aided drug design; consensus methods; fragment-based; machine learning; structure-based drug design.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. - DOI
-
- De Magalhães C.S., Almeida D.M., Barbosa H.J.C., Dardenne L.E. A dynamic niching genetic algorithm strategy for docking highly flexible ligands. Inf. Sci. 2014;289:206–224.
-
- De Magalhães C.S., Barbosa H.J.C., Dardenne L.E. Selection-Insertion Schemes in Genetic Algorithms for the Flexible Ligand Docking Problem. Lect. Notes Comput. Sci. 2004;3102:368–379.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

