Isothioureas, Ureas, and Their N-Methyl Amides from 2-Aminobenzothiazole and Chiral Amino Acids
- PMID: 31540462
- PMCID: PMC6767222
- DOI: 10.3390/molecules24183391
Isothioureas, Ureas, and Their N-Methyl Amides from 2-Aminobenzothiazole and Chiral Amino Acids
Abstract
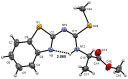
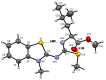
In this investigation, the reaction of 2-dithiomethylcarboimidatebenzothiazole with a series of six chiral amino-acids was studied. The reaction proceeds through the isolable sodium salt of SMe-isothiourea carboxylates as intermediates, whose reaction with methyl iodide in stirring DMF as solvent affords SMe-isothiourea methyl esters. The presence of water in the reaction leads to the corresponding urea carboxylates as isolable intermediates, whose methyl esters were obtained. Finally, the urea N-methyl amide derivatives were isolated when SMe-isothiourea or urea methyl esters were reacted with methylamine in the presence of water. The structures of synthesized compounds were established by 1H and 13C nuclear magnetic resonance and the structures of SMe-isothiourea methyl esters derived from (l)-glycine, (l)-alanine, (l)-phenylglycine, and (l)-leucine, by X-ray diffraction analysis. This methodology allows to functionalize 2-aminobenzothiazole with SMe-isothiourea, urea, and methylamide groups derived from chiral amino acids to get benzothiazole derivatives containing coordination sites and hydrogen bonding groups. Further research on the biological activities of some of these derivatives is ongoing.
Keywords: 2-aminobenzothiazole; 2-dithiomethylcarboimidatebenzothiazole; S-methyl-isothioureas; urea-carboxylate methyl esters and urea N-methyl amides; α-amino-acids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



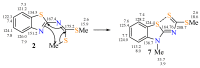




Similar articles
-
Non-racemic atropisomeric (thio)ureas as neutral enantioselective anion receptors for amino-acid derivatives: origin of smaller Kass with thiourea than urea derivatives.Chirality. 2006 Sep;18(9):762-71. doi: 10.1002/chir.20320. Chirality. 2006. PMID: 16871519
-
Anthracene derivatives bearing thiourea and glucopyranosyl groups for the highly selective chiral recognition of amino acids: opposite chiral selectivities from similar binding units.J Org Chem. 2008 Jan 4;73(1):301-4. doi: 10.1021/jo7022813. Epub 2007 Dec 6. J Org Chem. 2008. PMID: 18052393
-
Structure-enantioselectivity effects in 3,4-dihydropyrimido[2,1-b]benzothiazole-based isothioureas as enantioselective acylation catalysts.Org Biomol Chem. 2011 Jan 21;9(2):559-70. doi: 10.1039/c0ob00515k. Epub 2010 Nov 11. Org Biomol Chem. 2011. PMID: 21072411
-
Monoterpene-based chiral β-amino acid derivatives prepared from natural sources: syntheses and applications.Amino Acids. 2011 Aug;41(3):597-608. doi: 10.1007/s00726-011-0891-5. Epub 2011 Mar 30. Amino Acids. 2011. PMID: 21448657 Review.
-
Microbial/enzymatic synthesis of chiral drug intermediates.Adv Appl Microbiol. 2000;47:33-78. doi: 10.1016/s0065-2164(00)47001-2. Adv Appl Microbiol. 2000. PMID: 12876794 Review.
Cited by
-
In Silico and In Vivo Evaluation of Novel 2-Aminobenzothiazole Derivative Compounds as Antidiabetic Agents.Int J Mol Sci. 2025 Jan 22;26(3):909. doi: 10.3390/ijms26030909. Int J Mol Sci. 2025. PMID: 39940678 Free PMC article.
-
Synthesis and Biological Importance of 2-(thio)ureabenzothiazoles.Molecules. 2022 Sep 19;27(18):6104. doi: 10.3390/molecules27186104. Molecules. 2022. PMID: 36144837 Free PMC article. Review.
-
Preclinical Evaluation of 2-Aminobenzothiazole Derivatives: In Silico, In Vitro, and Preliminary In Vivo Studies as Diabetic Treatments and Their Complications.Molecules. 2025 Aug 20;30(16):3427. doi: 10.3390/molecules30163427. Molecules. 2025. PMID: 40871578 Free PMC article.
References
-
- Borthwick A.D., Davies D.E., Ertl P.F., Exall A.M., Haley T.M., Hart G.J., Jackson D.L., Parry N.R., Patikis A., Trivedi N., et al. Design and synthesis of pyrrolidine-5,5′-trans-lactams (5-oxo-hexahydropyrrolo [3, 2-b] pyrroles) as novel mechanism-based inhibitors of human cytomegalovirus protease. 4. Antiviral activity and plasma stability. J. Med. Chem. 2003;46:4428–4449. doi: 10.1021/jm030810w. - DOI - PubMed
-
- Saeed A., Rafique H., Hameed A., Rasheed S. Synthesis and antibacterial activity of some new 1-aroyl-3-(substituted-2-benzothiazolyl)thio-ureas. Pharm. Chem. J. 2008;42:191–195.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

