Glutathione Induced Immune-Stimulatory Activity by Promoting M1-Like Macrophages Polarization via Potential ROS Scavenging Capacity
- PMID: 31540482
- PMCID: PMC6770173
- DOI: 10.3390/antiox8090413
Glutathione Induced Immune-Stimulatory Activity by Promoting M1-Like Macrophages Polarization via Potential ROS Scavenging Capacity
Abstract
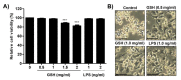
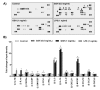
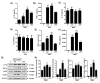
The present study investigated the immunomodulatory activity of reduced glutathione (GSH) by assessment of the macrophage polarization (MP)-mediated immune response in RAW 264.7 cells. Furthermore, we identified the signal pathway associated with immune regulation by GSH. The expressions of MP-associated cytokines and chemokines were assessed using cytokine array, nCounter Sprit platform, ELISA and immunoblotting. Phagocytosis activity and intracellular reactive oxygen species (ROS) generation were measured using fluorescence-activated cell sorter. As results of the cytokine array and nCounter gene array, GSH not only up-regulated pro-inflammatory cytokines, including interleukins and tumor necrosis factor-α, but also overexpressed neutrophil-attracting chemokines. Furthermore, GSH significantly stimulated the production of immune mediators, including nitric oxide and PGE2, as well as phagocytosis activity through nuclear factor kappa B activation. In addition, GSH significantly decreased LPS-induced ROS generation, which was associated with an activation of nuclear factor erythroid-derived 2-related factor 2 (Nrf2)/ heme oxygenease-1 (HO-1) signaling pathway. Our results suggest that GSH has potential ROS scavenging capacity via the induction of Nrf2-mediated HO-1, and immune-enhancing activity by regulation of M1-like macrophage polarization, indicating that GSH may be a useful strategy to increase the human defense system.
Keywords: antioxidant; glutathione; heme oxygenase-1; immune response; inflammatory cytokines; macrophage polarization; reactive oxygen species.
Conflict of interest statement
The authors have not any conflict of interest.
Figures





References
-
- Carrillo J.L.M., Garcia F.P.C., Coronado O.G., Garcia M.A.M., Cordero J.F.C. Physiology and pathology of innate immune response against pathogens. In: Rezaei N., editor. Physiology and Pathology of Immunology. IntechOpen Ltd.; London, UK: 2017. pp. 99–134. - DOI
-
- Van Rooijen N., Wijburg O.L., van den Dobbelsteen G.P., Sanders A. Macrophages in host defense mechanisms. Curr. Top. Microbiol. Immunol. 1996;210:159–165. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

