The control of gene expression and cell identity by H3K9 trimethylation
- PMID: 31540910
- PMCID: PMC6803365
- DOI: 10.1242/dev.181180
The control of gene expression and cell identity by H3K9 trimethylation
Abstract
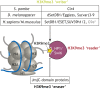
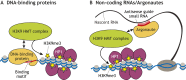
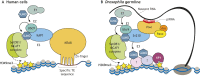
Histone 3 lysine 9 trimethylation (H3K9me3) is a conserved histone modification that is best known for its role in constitutive heterochromatin formation and the repression of repetitive DNA elements. More recently, it has become evident that H3K9me3 is also deposited at certain loci in a tissue-specific manner and plays important roles in regulating cell identity. Notably, H3K9me3 can repress genes encoding silencing factors, pointing to a fundamental principle of repressive chromatin auto-regulation. Interestingly, recent studies have shown that H3K9me3 deposition requires protein SUMOylation in different contexts, suggesting that the SUMO pathway functions as an important module in gene silencing and heterochromatin formation. In this Review, we discuss the role of H3K9me3 in gene regulation in various systems and the molecular mechanisms that guide the silencing machinery to target loci.
Keywords: Cell fate maintenance; Chromatin; Epigenetics; Gene regulation; Germline; Heterochromatin; Transcriptional repression; Transposons.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

References
-
- Aagaard L., Laible G., Selenko P., Schmid M., Dorn R., Schotta G., Kuhfittig S., Wolf A., Lebersorger A., Singh P. B. et al. (1999). Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 18, 1923-1938. 10.1093/emboj/18.7.1923 - DOI - PMC - PubMed
-
- Akkouche A., Mugat B., Barckmann B., Varela-Chavez C., Li B., Raffel R., Pélisson A. and Chambeyron S. (2017). Piwi is required during Drosophila embryogenesis to license dual-strand piRNA clusters for transposon repression in adult ovaries. Mol. Cell 66, 411-419.e4. 10.1016/j.molcel.2017.03.017 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

