Four Aromatic Intradiol Ring Cleavage Dioxygenases from Aspergillus niger
- PMID: 31540981
- PMCID: PMC6856329
- DOI: 10.1128/AEM.01786-19
Four Aromatic Intradiol Ring Cleavage Dioxygenases from Aspergillus niger
Abstract
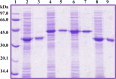
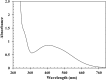
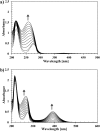
Ring cleavage dioxygenases catalyze the critical ring-opening step in the catabolism of aromatic compounds. The archetypal filamentous fungus Aspergillus niger previously has been reported to be able to utilize a range of monocyclic aromatic compounds as sole sources of carbon and energy. The genome of A. niger has been sequenced, and deduced amino acid sequences from a large number of gene models show various levels of similarity to bacterial intradiol ring cleavage dioxygenases, but no corresponding enzyme has been purified and characterized. Here, the cloning, heterologous expression, purification, and biochemical characterization of four nonheme iron(III)-containing intradiol dioxygenases (NRRL3_02644, NRRL3_04787, NRRL3_05330, and NRRL3_01405) from A. niger are reported. Purified enzymes were tested for their ability to cleave model catecholate substrates, and their apparent kinetic parameters were determined. Comparisons of kcat/Km values show that NRRL3_02644 and NRRL3_05330 are specific for hydroxyquinol (1,2,4-trihydroxybenzene), and phylogenetic analysis shows that these two enzymes are related to bacterial hydroxyquinol 1,2-dioxygenases. A high-activity catechol 1,2-dioxygenase (NRRL3_04787), which is phylogenetically related to other characterized and putative fungal catechol 1,2-dioxygenases, was also identified. The fourth enzyme (NRRL3_01405) appears to be a novel homodimeric Fe(III)-containing protocatechuate 3,4-dioxygenase that is phylogenetically distantly related to heterodimeric bacterial protocatechuate 3,4-dioxygenases. These investigations provide experimental evidence for the molecular function of these proteins and open the way to further investigations of the physiological roles for these enzymes in fungal metabolism of aromatic compounds.IMPORTANCE Aromatic ring opening using molecular oxygen is one of the critical steps in the degradation of aromatic compounds by microorganisms. While enzymes catalyzing this step have been well-studied in bacteria, their counterparts from fungi are poorly characterized despite the abundance of genes annotated as ring cleavage dioxygenases in fungal genomes. Aspergillus niger degrades a variety of aromatic compounds, and its genome harbors 5 genes encoding putative intracellular intradiol dioxygenases. The ability to predict the substrate specificities of the encoded enzymes from sequence data are limited. Here, we report the characterization of four purified intradiol ring cleavage dioxygenases from A. niger, revealing two hydroxyquinol-specific dioxygenases, a catechol dioxygenase, and a unique homodimeric protocatechuate dioxygenase. Their characteristics, as well as their phylogenetic relationships to predicted ring cleavage dioxygenases from other fungal species, provide insights into their molecular functions in aromatic compound metabolism by this fungus and other fungi.
Keywords: Aspergillus niger; aromatic compounds; dioxygenases.
Copyright © 2019 American Society for Microbiology.
Figures

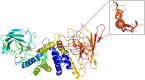


Similar articles
-
Cloning of a gene encoding hydroxyquinol 1,2-dioxygenase that catalyzes both intradiol and extradiol ring cleavage of catechol.Biosci Biotechnol Biochem. 1999 May;63(5):859-65. doi: 10.1271/bbb.63.859. Biosci Biotechnol Biochem. 1999. PMID: 10380628
-
Functional analysis of the protocatechuate branch of the β-ketoadipate pathway in Aspergillus niger.J Biol Chem. 2023 Aug;299(8):105003. doi: 10.1016/j.jbc.2023.105003. Epub 2023 Jul 1. J Biol Chem. 2023. PMID: 37399977 Free PMC article.
-
Bacterial aromatic ring-cleavage enzymes are classified into two different gene families.J Biol Chem. 1989 Sep 15;264(26):15328-33. J Biol Chem. 1989. PMID: 2670937
-
The ins and outs of ring-cleaving dioxygenases.Crit Rev Biochem Mol Biol. 2006 Jul-Aug;41(4):241-67. doi: 10.1080/10409230600817422. Crit Rev Biochem Mol Biol. 2006. PMID: 16849108 Review.
-
Biophysical analyses of designed and selected mutants of protocatechuate 3,4-dioxygenase1.Annu Rev Microbiol. 2004;58:555-85. doi: 10.1146/annurev.micro.57.030502.090927. Annu Rev Microbiol. 2004. PMID: 15487948 Review.
Cited by
-
Identification of a Conserved Transcriptional Activator-Repressor Module Controlling the Expression of Genes Involved in Tannic Acid Degradation and Gallic Acid Utilization in Aspergillus niger.Front Fungal Biol. 2021 May 25;2:681631. doi: 10.3389/ffunb.2021.681631. eCollection 2021. Front Fungal Biol. 2021. PMID: 37744122 Free PMC article.
-
Twists and Turns in the Salicylate Catabolism of Aspergillus terreus, Revealing New Roles of the 3-Hydroxyanthranilate Pathway.mSystems. 2021 Jan 26;6(1):e00230-20. doi: 10.1128/mSystems.00230-20. mSystems. 2021. PMID: 33500329 Free PMC article.
-
Discovery and Functional Analysis of a Salicylic Acid Hydroxylase from Aspergillus niger.Appl Environ Microbiol. 2021 Feb 26;87(6):e02701-20. doi: 10.1128/AEM.02701-20. Print 2021 Feb 26. Appl Environ Microbiol. 2021. PMID: 33397706 Free PMC article.
-
Transcriptome analysis of Aspergillus oryzae RIB40 under chemical stress reveals mechanisms of adaptation to fungistatic compounds of lignocellulosic side streams.Biotechnol Biofuels Bioprod. 2025 Aug 8;18(1):89. doi: 10.1186/s13068-025-02688-5. Biotechnol Biofuels Bioprod. 2025. PMID: 40781683 Free PMC article.
-
Characterization of two 1,2,4-trihydroxybenzene 1,2-dioxygenases from Phanerochaete chrysosporium.Appl Microbiol Biotechnol. 2022 Jun;106(12):4499-4509. doi: 10.1007/s00253-022-12007-9. Epub 2022 Jun 10. Appl Microbiol Biotechnol. 2022. PMID: 35687156
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

